Combined use of eDNA metabarcoding and video surveillance for the assessment of fish biodiversity
- PMID: 30004598
- PMCID: PMC7379492
- DOI: 10.1111/cobi.13183
Combined use of eDNA metabarcoding and video surveillance for the assessment of fish biodiversity
Abstract
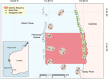
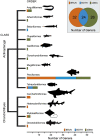
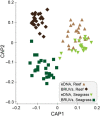
Monitoring communities of fish is important for the management and sustainability of fisheries and marine ecosystems. Baited remote underwater video systems (BRUVs) are among the most effective nondestructive techniques for sampling bony fishes and elasmobranchs (sharks, rays, and skates). However, BRUVs sample visually conspicuous biota; hence, some taxa are undersampled or not recorded at all. We compared the diversity of fishes characterized using BRUVs with diversity detected via environmental DNA (eDNA) metabarcoding. We sampled seawater and captured BRUVs imagery at 48 locales that included reef and seagrass beds inside and outside a marine reserve (Jurien Bay in Western Australia). Eighty-two fish genera from 13 orders were detected, and the community of fishes described using eDNA and BRUVs combined yielded >30% more generic richness than when either method was used alone. Rather than detecting a homogenous genetic signature, the eDNA assemblages mirrored the BRUVs' spatial explicitness; differentiation of taxa between seagrass and reef was clear despite the relatively small geographical scale of the study site (∼35 km2 ). Taxa that were not sampled by one approach, due to limitations and biases intrinsic to the method, were often detected with the other. Therefore, using BRUVs and eDNA in concert provides a more holistic view of vertebrate marine communities across habitats. Both methods are noninvasive, which enhances their potential for widespread implementation in the surveillance of marine ecosystems.
Uso Combinado del Metacódigo de Barras de eDNA y Videograbaciones para la Evaluación de la Biodiversidad de Peces
Resumen: El monitoreo de comunidades de peces es importante para el manejo y sustentabilidad de las pesquerías y los ecosistemas marinos. Los sistemas remotos de video submarino con carnada (SRVSC) están entre las técnicas no destructivas más efectivas para el muestreo de peces óseos y elasmobranquios (tiburones, mantarrayas y rayas). Sin embargo, los SRVSC muestrean biota que es conspicua visiblemente; entonces, algunos taxones están mal muestreados o simplemente no se registran en los muestreos. Comparamos la diversidad de peces caracterizada usando SRVSC con la diversidad detectada por medio del metacódigo de barras de ADN ambiental (eDNA, en inglés). Muestreamos el agua de mar y capturamos imágenes con SRVSC en 48 localidades que incluyeron el arrecife y los pastos marinos dentro y fuera de una reserva marina (Bahía Jurien en el oeste de Australia). Se detectaron 83 géneros de peces de 13 órdenes, y la comunidad de peces descrita con el uso combinado del eDNA y el SRVSC produjo >30% riqueza más genérica que cuando cualquiera de los dos métodos se usó individualmente. En lugar de detectar una firma genética homogénea, los ensamblados de eDNA reflejaron la claridad espacial del SRVSC; la diferenciación de los taxones entre los pastos marinos y el arrecife fue clara a pesar la escala geográfica relativamente pequeña del sitio de estudio (∼35 km2). Los taxones que no fueron muestreados por uno de los métodos, por causa de limitaciones y sesgos intrínsecos al método, casi siempre fueron detectados usando el otro método. Por lo tanto, el uso de SRVSC y el eDNA en concreto proporciona una visión más holística de las comunidades marinas de vertebrados en todos los hábitats. Ambos métodos son no invasivos, lo que incrementa su potencial para ser una implementación de uso amplio en la vigilancia de los ecosistemas marinos.
Keywords: ADN ambiental; baited remote underwater video systems; elasmobranchs; elasmobranquios; environmental DNA; environmental genomics; genómica ambiental; manejo marino; marine management; sistemas remotos de video submarino con carnada.
© 2018 The Authors. Conservation Biology published by Wiley Periodicals, Inc.
Figures
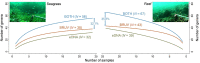
References
-
- Alberdi A, Aizpurua O, Gilbert MT, Bohmann K. 2017. Scrutinizing key steps for reliable metabarcoding of environmental samples. Methods in Ecology and Evolution 2018:134–147.
-
- Anderson MJ, Gorley RN, Clarke KR. 2008. PERMANOVA+ for primer: guide to software and statistical methods. Plymouth, United Kingdom.
-
- Barnes MA, Turner CR. 2016. The ecology of environmental DNA and implications for conservation genetics. Conservation Genetics 17:1–17.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

