Decoding the chromatin proteome of a single genomic locus by DNA sequencing
- PMID: 30005073
- PMCID: PMC6059479
- DOI: 10.1371/journal.pbio.2005542
Decoding the chromatin proteome of a single genomic locus by DNA sequencing
Abstract
Transcription, replication, and repair involve interactions of specific genomic loci with many different proteins. How these interactions are orchestrated at any given location and under changing cellular conditions is largely unknown because systematically measuring protein-DNA interactions at a specific locus in the genome is challenging. To address this problem, we developed Epi-Decoder, a Tag-chromatin immunoprecipitation-Barcode-Sequencing (TAG-ChIP-Barcode-Seq) technology in budding yeast. Epi-Decoder is orthogonal to proteomics approaches because it does not rely on mass spectrometry (MS) but instead takes advantage of DNA sequencing. Analysis of the proteome of a transcribed locus proximal to an origin of replication revealed more than 400 interacting proteins. Moreover, replication stress induced changes in local chromatin proteome composition prior to local origin firing, affecting replication proteins as well as transcription proteins. Finally, we show that native genomic loci can be decoded by efficient construction of barcode libraries assisted by clustered regularly interspaced short palindromic repeats and CRISPR-associated protein 9 (CRISPR/Cas9). Thus, Epi-Decoder is an effective strategy to identify and quantify in an unbiased and systematic manner the proteome of an individual genomic locus by DNA sequencing.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

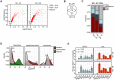

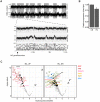
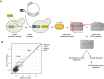
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

