Heterologous Expression of Pseudomonas putida Methyl-Accepting Chemotaxis Proteins Yields Escherichia coli Cells Chemotactic to Aromatic Compounds
- PMID: 30006400
- PMCID: PMC6121982
- DOI: 10.1128/AEM.01362-18
Heterologous Expression of Pseudomonas putida Methyl-Accepting Chemotaxis Proteins Yields Escherichia coli Cells Chemotactic to Aromatic Compounds
Abstract
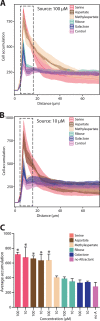
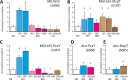
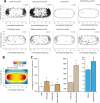
Escherichia coli, commonly used in chemotaxis studies, is attracted mostly by amino acids, sugars, and peptides. We envisioned modifying the chemotaxis specificity of E. coli by expressing heterologous chemoreceptors from Pseudomonas putida enabling attraction either to toluene or benzoate. The mcpT gene encoding the type 40-helical bundle (40H) methyl-accepting chemoreceptor for toluene from Pseudomonas putida MT53 and the pcaY gene for the type 40H receptor for benzoate and related molecules from P. putida F1 were expressed from the trg promoter on a plasmid in motile wild-type E. coli MG1655. E. coli cells expressing McpT accumulated in chemoattraction assays to sources with 60 to 200 μM toluene, although less strongly than the response to 100 μM serine, but statistically significantly stronger than that to sources without any added attractant. An McpT-mCherry fusion protein was detectably expressed in E. coli and yielded weak but distinguishable membranes and polar foci in 1% of cells. E. coli cells expressing PcaY showed weak attraction to 0.1 to 1 mM benzoate, but 50 to 70% of cells localized the PcaY-mCherry fusion to their membrane. We conclude that implementing heterologous receptors in the E. coli chemotaxis network is possible and, upon improvement of the compatibility of the type 40H chemoreceptors, may bear interest for biosensing.IMPORTANCE Bacterial chemotaxis might be harnessed for the development of rapid biosensors, in which chemical availability is deduced from cell accumulation to chemoattractants over time. Chemotaxis of Escherichia coli has been well studied, but the bacterium is not attracted to chemicals of environmental concern, such as aromatic solvents. We show here that heterologous chemoreceptors for aromatic compounds from Pseudomonas putida at least partly functionally complement the E. coli chemotaxis network, yielding cells attracted to toluene or benzoate. Complementation was still inferior to native chemoattractants, like serine, but our study demonstrates the potential for obtaining selective sensing for aromatic compounds in E. coli.
Keywords: biosensing; chemotaxis.
Copyright © 2018 American Society for Microbiology.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

