MR1 displays the microbial metabolome driving selective MR1-restricted T cell receptor usage
- PMID: 30006464
- PMCID: PMC7085347
- DOI: 10.1126/sciimmunol.aao2556
MR1 displays the microbial metabolome driving selective MR1-restricted T cell receptor usage
Abstract
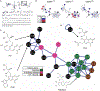
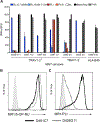
MR1-restricted T cells (MR1Ts) are a T cell subset that recognize and mediate host defense to a broad array of microbial pathogens, including respiratory pathogens (e.g., Mycobacterium tuberculosis, Streptococcus pyogenes, and Francisella tularensis) and enteric pathogens (e.g., Escherichia coli and Salmonella species). Mucosal-associated invariant T (MAIT) cells, a subset of MR1Ts, were historically defined by the use of a semi-invariant T cell receptor (TCR) and recognition of small molecules derived from the riboflavin biosynthesis pathway presented on MR1. We used mass spectrometry to identify the repertoire of ligands presented by MR1 from the microbes E. coli and Mycobacterium smegmatis We found that the MR1 ligandome is unexpectedly broad, revealing functionally distinct ligands derived from E. coli and M. smegmatis The identification, synthesis, and functional analysis of mycobacterial ligands reveal that MR1T ligands can be distinguished by MR1Ts with diverse TCR usage. These data demonstrate that MR1 can serve as an immune sensor of the microbial ligandome.
Copyright © 2018 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Competing interests:
The authors declare that they have no competing interests.
Figures



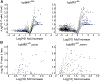

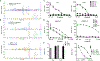
References
-
- Riegert P, Wanner V, Bahram S, Genomics, isoforms, expression, and phylogeny of the MHC class I-related MR1 gene. J. Immunol 161, 4066–4077 (1998). - PubMed
-
- Gold MC, Cerri S, Smyk-Pearson S, Cansler ME, Vogt TM, Delepine J, Winata E, Swarbrick GM, Chua WJ, Yu YYL, Lantz O, Cook MS, Null MD, Jacoby DB, Harriff MJ, Lewinsohn DA, Hansen TH, Lewinsohn DM, Human mucosal associated invariant T cells detect bacterially infected cells. PLOS Biol. 8, e1000407 (2010). - PMC - PubMed
-
- Le Bourhis L, Martin E, Péguillet I, Guihot A, Froux N, Coré M, Lévy E, Dusseaux M, Meyssonnier V, Premel V, Ngo C, Riteau B, Duban L, Robert D, Huang S, Rottman M, Soudais C, Lantz O, Antimicrobial activity of mucosal-associated invariant T cells. Nat. Immunol 11, 701–708 (2010). - PubMed
-
- Dusseaux M, Martin E, Serriari N, Péguillet I, Premel V, Louis D, Milder M, Le Bourhis L, Soudais C, Treiner E, Lantz O, Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood 117, 1250–1259 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

