Universal Plant Phosphoproteomics Workflow and Its Application to Tomato Signaling in Response to Cold Stress
- PMID: 30006488
- PMCID: PMC6166681
- DOI: 10.1074/mcp.TIR118.000702
Universal Plant Phosphoproteomics Workflow and Its Application to Tomato Signaling in Response to Cold Stress
Abstract
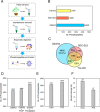
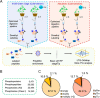
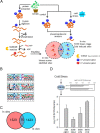
Phosphorylation-mediated signaling transduction plays a crucial role in the regulation of plant defense mechanisms against environmental stresses. To address the high complexity and dynamic range of plant proteomes and phosphoproteomes, we present a universal sample preparation procedure that facilitates plant phosphoproteomic profiling. This advanced workflow significantly improves phosphopeptide identifications, enabling deep insight into plant phosphoproteomes. We then applied the workflow to study the phosphorylation events involved in tomato cold tolerance mechanisms. Phosphoproteomic changes of two tomato species (N135 Green Gage and Atacames) with distinct cold tolerance phenotypes were profiled under cold stress. In total, we identified more than 30,000 unique phosphopeptides from tomato leaves, representing about 5500 phosphoproteins, thereby creating the largest tomato phosphoproteomic resource to date. The data, along with the validation through in vitro kinase reactions, allowed us to identify kinases involved in cold tolerant signaling and discover distinctive kinase-substrate events in two tomato species in response to a cold environment. The activation of SnRK2s and their direct substrates may assist N135 Green Gage tomatoes in surviving long-term cold stress. Taken together, the streamlined approach and the resulting deep phosphoproteomic analyses revealed a global view of tomato cold-induced signaling mechanisms.
Keywords: Cold Stress; Mass Spectrometry; Phosphoproteome; Phosphorylation; Signal Transduction; Stress response; Tomato Phosphoproteome.
© 2018 Hsu et al.
Conflict of interest statement
The authors have declared no conflict of interest
Figures

References
-
- Schulze W. X. (2010) Proteomics approaches to understand protein phosphorylation in pathway modulation. Curr. Opin. Plant Biol. 13, 280–287 - PubMed
-
- Silva-Sanchez C., Li H., and Chen S. (2015) Recent advances and challenges in plant phosphoproteomics. Proteomics 15, 1127–1141 - PubMed
-
- Singh D. K., Calvino M., Brauer E. K., Fernandez-Pozo N., Strickler S., Yalamanchili R., Suzuki H., Aoki K., Shibata D., Stratmann J. W., Popescu G. V., Mueller L. A., and Popescu S. C. (2014) The tomato kinome and the tomato kinase library ORFeome: novel resources for the study of kinases and signal transduction in tomato and solanaceae species. Mol. Plant Microbe Interact. 27, 7–17 - PubMed
-
- Chinnusamy V., Zhu J., and Zhu J. K. (2007) Cold stress regulation of gene expression in plants. Trends Plant Sci. 12, 444–451 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

