Structure and energy based quantitative missense variant effect analysis provides insights into drug resistance mechanisms of anaplastic lymphoma kinase mutations
- PMID: 30006516
- PMCID: PMC6045602
- DOI: 10.1038/s41598-018-28752-9
Structure and energy based quantitative missense variant effect analysis provides insights into drug resistance mechanisms of anaplastic lymphoma kinase mutations
Abstract
Anaplastic lymphoma kinase (ALK) is considered as a validated molecular target in multiple malignancies, such as non-small cell lung cancer (NSCLC). However, the effectiveness of molecularly targeted therapies using ALK inhibitors is almost universally limited by drug resistance. Drug resistance to molecularly targeted therapies has now become a major obstacle to effective cancer treatment and personalized medicine. It is of particular importance to provide an improved understanding on the mechanisms of resistance of ALK inhibitors, thus rational new therapeutic strategies can be developed to combat resistance. We used state-of-the-art computational approaches to systematically explore the mutational effects of ALK mutations on drug resistance properties. We found the activation of ALK was increased by substitution with destabilizing mutations, creating the capacity to confer drug resistance to inhibitors. In addition, results implied that evolutionary constraints might affect the drug resistance properties. Moreover, an extensive profile of drugs against ALK mutations was constructed to give better understanding of the mechanism of drug resistance based on structural transitions and energetic variation. Our work hopes to provide an up-to-date mechanistic framework for understanding the mechanisms of drug resistance induced by ALK mutations, thus tailor treatment decisions after the emergence of resistance in ALK-dependent diseases.
Conflict of interest statement
The authors declare no competing interests.
Figures








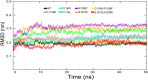

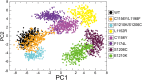

Similar articles
-
Resistance Mechanisms to Targeted Therapies in ROS1+ and ALK+ Non-small Cell Lung Cancer.Clin Cancer Res. 2018 Jul 15;24(14):3334-3347. doi: 10.1158/1078-0432.CCR-17-2452. Epub 2018 Apr 10. Clin Cancer Res. 2018. PMID: 29636358 Free PMC article.
-
Acquired Resistance Mutations to ALK Inhibitors Identified by Single Circulating Tumor Cell Sequencing in ALK-Rearranged Non-Small-Cell Lung Cancer.Clin Cancer Res. 2019 Nov 15;25(22):6671-6682. doi: 10.1158/1078-0432.CCR-19-1176. Epub 2019 Aug 22. Clin Cancer Res. 2019. PMID: 31439588
-
Next generation sequencing reveals a novel ALK G1128A mutation resistant to crizotinib in an ALK-Rearranged NSCLC patient.Lung Cancer. 2018 Sep;123:83-86. doi: 10.1016/j.lungcan.2018.07.004. Epub 2018 Jul 6. Lung Cancer. 2018. PMID: 30089600
-
Management of Resistance to First-Line Anaplastic Lymphoma Kinase Tyrosine Kinase Inhibitor Therapy.Curr Treat Options Oncol. 2018 May 28;19(7):37. doi: 10.1007/s11864-018-0553-x. Curr Treat Options Oncol. 2018. PMID: 29808239 Review.
-
Resistance to Crizotinib in Advanced Non-Small Cell Lung Cancer (NSCLC) with ALK Rearrangement: Mechanisms, Treatment Strategies and New Targeted Therapies.Curr Clin Pharmacol. 2016;11(2):77-87. doi: 10.2174/1574884711666160502124134. Curr Clin Pharmacol. 2016. PMID: 27138017 Review.
Cited by
-
Targeting ALK Rearrangements in NSCLC: Current State of the Art.Front Oncol. 2022 Apr 6;12:863461. doi: 10.3389/fonc.2022.863461. eCollection 2022. Front Oncol. 2022. PMID: 35463328 Free PMC article. Review.
-
Deciphering the Mechanism of Gilteritinib Overcoming Lorlatinib Resistance to the Double Mutant I1171N/F1174I in Anaplastic Lymphoma Kinase.Front Cell Dev Biol. 2021 Dec 23;9:808864. doi: 10.3389/fcell.2021.808864. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 35004700 Free PMC article.
-
Next-generation sequencing to dynamically detect mechanisms of resistance to ALK inhibitors in ALK-positive NSCLC patients: a case report.Transl Lung Cancer Res. 2020 Apr;9(2):366-372. doi: 10.21037/tlcr.2020.02.07. Transl Lung Cancer Res. 2020. PMID: 32420077 Free PMC article.
-
Resistance mutations and the blood-brain barrier: Key challenges in targeted treatment of brain metastatic non-small cell lung cancer.Acta Pharm Sin B. 2025 Aug;15(8):3833-3851. doi: 10.1016/j.apsb.2025.06.002. Epub 2025 Jun 7. Acta Pharm Sin B. 2025. PMID: 40893689 Free PMC article. Review.
-
SNP2SIM: a modular workflow for standardizing molecular simulation and functional analysis of protein variants.BMC Bioinformatics. 2019 Apr 3;20(1):171. doi: 10.1186/s12859-019-2774-9. BMC Bioinformatics. 2019. PMID: 30943891 Free PMC article.
References
-
- Morris, S. W. et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science-New York Then Washington-, 1281–1281 (1994). - PubMed
-
- Shiota M, et al. Hyperphosphorylation of a novel 80 kDa protein-tyrosine kinase similar to Ltk in a human Ki-1 lymphoma cell line, AMS3. Oncogene. 1994;9:1567–1574. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 81373311/National Natural Science Foundation of China (National Science Foundation of China)/International
- 31300674/National Natural Science Foundation of China (National Science Foundation of China)/International
- 81173093/National Natural Science Foundation of China (National Science Foundation of China)/International
- J1103518/National Natural Science Foundation of China (National Science Foundation of China)/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

