Autophagy as a promoter of longevity: insights from model organisms
- PMID: 30006559
- PMCID: PMC6424591
- DOI: 10.1038/s41580-018-0033-y
Autophagy as a promoter of longevity: insights from model organisms
Erratum in
-
Publisher Correction: Autophagy as a promoter of longevity: insights from model organisms.Nat Rev Mol Cell Biol. 2018 Sep;19(9):611. doi: 10.1038/s41580-018-0048-4. Nat Rev Mol Cell Biol. 2018. PMID: 30046055
Abstract
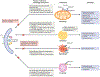
Autophagy is a conserved process that catabolizes intracellular components to maintain energy homeostasis and to protect cells against stress. Autophagy has crucial roles during development and disease, and evidence accumulated over the past decade indicates that autophagy also has a direct role in modulating ageing. In particular, elegant studies using yeasts, worms, flies and mice have demonstrated a broad requirement for autophagy-related genes in the lifespan extension observed in a number of conserved longevity paradigms. Moreover, several new and interesting concepts relevant to autophagy and its role in modulating longevity have emerged. First, select tissues may require or benefit from autophagy activation in longevity paradigms, as tissue-specific overexpression of single autophagy genes is sufficient to extend lifespan. Second, selective types of autophagy may be crucial for longevity by specifically targeting dysfunctional cellular components and preventing their accumulation. And third, autophagy can influence organismal health and ageing even non-cell autonomously, and thus, autophagy stimulation in select tissues can have beneficial, systemic effects on lifespan. Understanding these mechanisms will be important for the development of approaches to improve human healthspan that are based on the modulation of autophagy.
Conflict of interest statement
Competing interests
D.C.R. is a consultant for E3Bio and has consulted for GlaxoSmithKline and AstraZeneca. D.C.R. has grant support from AstraZeneca and AbbVie.
Figures
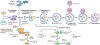
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

