ATP synthase F1 subunits recruited to centromeres by CENP-A are required for male meiosis
- PMID: 30006572
- PMCID: PMC6045659
- DOI: 10.1038/s41467-018-05093-9
ATP synthase F1 subunits recruited to centromeres by CENP-A are required for male meiosis
Abstract
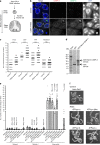
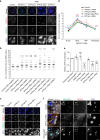
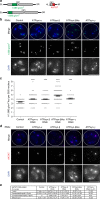
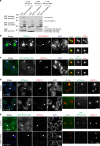
The histone H3 variant CENP-A epigenetically defines the centromere and is critical for chromosome segregation. Here we report an interaction between CENP-A and subunits of the mitochondrial ATP synthase complex in the germline of male Drosophila. Furthermore, we report that knockdown of CENP-A, as well as subunits ATPsyn-α, -βlike (a testis-specific paralogue of ATPsyn-β) and -γ disrupts sister centromere cohesion in meiotic prophase I. We find that this disruption is likely independent of reduced ATP levels. We identify that ATPsyn-α and -βlike localise to meiotic centromeres and that this localisation is dependent on the presence of CENP-A. We show that ATPsyn-α directly interacts with the N-terminus of CENP-A in vitro and that truncation of its N terminus perturbs sister centromere cohesion in prophase I. We propose that the CENP-A N-terminus recruits ATPsyn-α and -βlike to centromeres to promote sister centromere cohesion in a nuclear function that is independent of oxidative phosphorylation.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

