SecA inhibitors as potential antimicrobial agents: differential actions on SecA-only and SecA-SecYEG protein-conducting channels
- PMID: 30007321
- PMCID: PMC7190897
- DOI: 10.1093/femsle/fny145
SecA inhibitors as potential antimicrobial agents: differential actions on SecA-only and SecA-SecYEG protein-conducting channels
Abstract
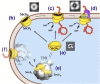
Sec-dependent protein translocation is an essential process in bacteria. SecA is a key component of the translocation machinery and has multiple domains that interact with various ligands. SecA acts as an ATPase motor to drive the precursor protein/peptide through the SecYEG protein translocation channels. As SecA is unique to bacteria and there is no mammalian counterpart, it is an ideal target for the development of new antimicrobials. Several reviews detail the assays for ATPase and protein translocation, as well as the search for SecA inhibitors. Recent studies have shown that, in addition to the SecA-SecYEG translocation channels, there are SecA-only channels in the lipid bilayers, which function independently from the SecYEG machinery. This mini-review focuses on recent advances on the newly developed SecA inhibitors that allow the evaluation of their potential as antimicrobial agents, as well as a fundamental understanding of mechanisms of SecA function(s). These SecA inhibitors abrogate the effects of efflux pumps in both Gram-positive and Gram-negative bacteria. We also discuss recent findings that SecA binds to ribosomes and nascent peptides, which suggest other roles of SecA. A model for the multiple roles of SecA is presented.
Figures

References
-
- Akula N, Zheng H, Han FQ et al. . Discovery of novel SecA inhibitors of Candidatus Liberibacter asiaticus by structure based design. Bioorg Med Chem Lett 2011;21:4183–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

