DNA binding with a minimal scaffold: structure-function analysis of Lig E DNA ligases
- PMID: 30007325
- PMCID: PMC6144786
- DOI: 10.1093/nar/gky622
DNA binding with a minimal scaffold: structure-function analysis of Lig E DNA ligases
Abstract
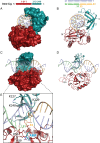
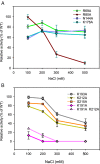
DNA ligases join breaks in the phosphodiester backbone of DNA by catalysing the formation of bonds between opposing 5'P and 3'OH ends in an adenylation-dependent manner. Catalysis is accompanied by reorientation of two core domains to provide access to the active site for cofactor utilization and enable substrate binding and product release. The general paradigm is that DNA ligases engage their DNA substrate through complete encirclement of the duplex, completed by inter-domain kissing contacts via loops or additional domains. The recent structure of a minimal Lig E-type DNA ligase, however, implies it must use a different mechanism, as it lacks any domains or loops appending the catalytic core which could complete encirclement. In the present study, we have used a structure-guided mutagenesis approach to investigate the role of conserved regions in the Lig E proteins with respect to DNA binding. We report the structure of a Lig-E type DNA ligase bound to the nicked DNA-adenylate reaction intermediate, confirming that complete encirclement is unnecessary for substrate engagement. Biochemical and biophysical measurements of point mutants to residues implicated in binding highlight the importance of basic residues in the OB domain, and inter-domain contacts to the linker.
Figures

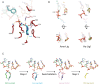



References
-
- Tomkinson A.E., Vijayakumar S., Pascal J.M., Ellenberger T.. DNA ligases: structure, reaction mechanism, and function. Chem. Rev. 2006; 106:687–699. - PubMed
-
- Nair P.A., Nandakumar J., Smith P., Odell M., Lima C.D., Shuman S.. Structural basis for nick recognition by a minimal pluripotent DNA ligase. Nat. Struct. Mol. Biol. 2007; 14:770–778. - PubMed
-
- Pitcher R.S., Brissett N.C., Doherty A.J.. Nonhomologous end-joining in bacteria: a microbial perspective. Annu. Rev. Microbiol. 2007; 61:259–282. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

