Transcriptome reprogramming by cancer exosomes: identification of novel molecular targets in matrix and immune modulation
- PMID: 30008265
- PMCID: PMC6047127
- DOI: 10.1186/s12943-018-0846-5
Transcriptome reprogramming by cancer exosomes: identification of novel molecular targets in matrix and immune modulation
Abstract
Background: Exosomes are extracellular vesicles released by almost all cell types, including cancer cells, into bodily fluids such as saliva, plasma, breast milk, semen, urine, cerebrospinal fluid, amniotic fluid, synovial fluid and sputum. Their key function being intercellular communication with both neighbouring as well as distant cells. Cancer exosomes have been shown to regulate organ-specific metastasis. However, little is known about the functional differences and molecular consequences of normal cells responding to exosomes derived from normal cells compared to those derived from cancer cells.
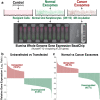
Methods: Here, we characterised and compared the transcriptome profiles of primary human normal oral keratinocytes (HNOK) in response to exosomes isolated from either primary HNOK or head and neck squamous cell carcinoma (HNSCC) cell lines.
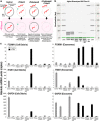
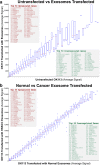
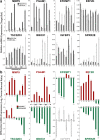
Results: In recipient HNOK cells, we found that regardless of normal or cancer derived, exosomes altered molecular programmes involved in matrix modulation (MMP9), cytoskeletal remodelling (TUBB6, FEZ1, CCT6A), viral/dsRNA-induced interferon (OAS1, IFI6), anti-inflammatory (TSC22D3), deubiquitin (OTUD1), lipid metabolism and membrane trafficking (BBOX1, LRP11, RAB6A). Interestingly, cancer exosomes, but not normal exosomes, modulated expression of matrix remodelling (EFEMP1, DDK3, SPARC), cell cycle (EEF2K), membrane remodelling (LAMP2, SRPX), differentiation (SPRR2E), apoptosis (CTSC), transcription/translation (KLF6, PUS7). We have also identified CEP55 as a potential cancer exosomal marker.
Conclusions: In conclusion, both normal and cancer exosomes modulated unique gene expression pathways in normal recipient cells. Cancer cells may exploit exosomes to confer transcriptome reprogramming that leads to cancer-associated pathologies such as angiogenesis, immune evasion/modulation, cell fate alteration and metastasis. Molecular pathways and biomarkers identified in this study may be clinically exploitable for developing novel liquid-biopsy based diagnostics and immunotherapies.
Keywords: Biomarkers; CEP55; ESCRT, exosomes; Extracellular vesicles; FOXM1; Reprogramming.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

