Strain-Specific Contribution of Eukaryotic Elongation Factor 1 Gamma to the Translation of Influenza A Virus Proteins
- PMID: 30008712
- PMCID: PMC6033995
- DOI: 10.3389/fmicb.2018.01446
Strain-Specific Contribution of Eukaryotic Elongation Factor 1 Gamma to the Translation of Influenza A Virus Proteins
Abstract
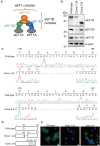
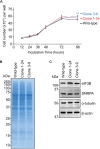
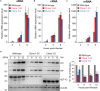
Influenza A virus exploits multiple host proteins during infection. To define the virus-host interactome, our group conducted a proteomics-based screen and identified 299 genes that contributed to virus replication and 24 genes that were antiviral. Of these genes, we focused on the role during virus replication of eukaryotic elongation factor 1 gamma (eEF1G), which is a subunit of the eukaryotic elongation factor-1 complex and known to be a pro-viral host protein. Using the CRISPR/Cas9 system, we obtained two clones that were defective in eEF1G expression. In both of these clones, A/WSN/33 (H1N1) virus growth and protein expression were significantly suppressed, but viral mRNA, vRNA, and cRNA expression were not reduced. However, the replication and protein expression of A/California/04/2009 (H1N1pdm) virus in both clones were similar to those in parental cells. We found that the PB2 and PA proteins of WSN virus were responsible for the eEF1G-dependent replication. Our data show that eEF1G plays a role in the translation of virus proteins in a strain-specific manner. Additional analyses may be needed to further understand the role of strain-specific host proteins during virus replication.
Keywords: PA; PB2; eEF1G; host protein; influenza virus; protein translation.
Figures



Similar articles
-
eEF1G interaction with foot-and-mouth disease virus nonstructural protein 2B: Identification by yeast two-hybrid system.Microb Pathog. 2017 Nov;112:111-116. doi: 10.1016/j.micpath.2017.09.039. Epub 2017 Sep 21. Microb Pathog. 2017. PMID: 28942178
-
Inhibition of Avian Influenza A Virus Replication in Human Cells by Host Restriction Factor TUFM Is Correlated with Autophagy.mBio. 2017 Jun 13;8(3):e00481-17. doi: 10.1128/mBio.00481-17. mBio. 2017. PMID: 28611246 Free PMC article.
-
Influenza A Virus Protein PA-X Contributes to Viral Growth and Suppression of the Host Antiviral and Immune Responses.J Virol. 2015 Jun;89(12):6442-52. doi: 10.1128/JVI.00319-15. Epub 2015 Apr 8. J Virol. 2015. PMID: 25855745 Free PMC article.
-
[Transcription and replication of influenza virus genome].Nihon Rinsho. 1997 Oct;55(10):2555-61. Nihon Rinsho. 1997. PMID: 9360371 Review. Japanese.
-
The molecular anatomy of influenza virus RNA polymerase.Biol Chem. 1997 Jun;378(6):483-8. Biol Chem. 1997. PMID: 9224927 Review.
Cited by
-
Leishmania infection upregulates and engages host macrophage Argonaute 1, and system-wide proteomics reveals Argonaute 1-dependent host response.Front Immunol. 2023 Nov 30;14:1287539. doi: 10.3389/fimmu.2023.1287539. eCollection 2023. Front Immunol. 2023. PMID: 38098491 Free PMC article.
-
Plitidepsin: Mechanisms and Clinical Profile of a Promising Antiviral Agent against COVID-19.J Pers Med. 2021 Jul 16;11(7):668. doi: 10.3390/jpm11070668. J Pers Med. 2021. PMID: 34357135 Free PMC article. Review.
-
Multidimensional Regulatory Mechanisms and Targeting Strategies of the eEF1 Family in RNA Virus Infection.Viruses. 2025 May 7;17(5):682. doi: 10.3390/v17050682. Viruses. 2025. PMID: 40431694 Free PMC article. Review.
-
Clinically Evaluated COVID-19 Drugs with Therapeutic Potential for Biological Warfare Agents.Microorganisms. 2023 Jun 14;11(6):1577. doi: 10.3390/microorganisms11061577. Microorganisms. 2023. PMID: 37375079 Free PMC article. Review.
-
Eukaryotic Translation Elongation Factor 1 Delta Inhibits the Nuclear Import of the Nucleoprotein and PA-PB1 Heterodimer of Influenza A Virus.J Virol. 2020 Dec 22;95(2):e01391-20. doi: 10.1128/JVI.01391-20. Print 2020 Dec 22. J Virol. 2020. PMID: 33087462 Free PMC article.
References
-
- Gillen C. M., Gao Y., Niehaus-Sauter M. M., Wylde M. R., Wheatly M. G. (2008). Elongation factor 1Bgamma (eEF1Bgamma) expression during the molting cycle and cold acclimation in the crayfish Procambarus clarkii. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 150 170–176. 10.1016/j.cbpb.2008.02.010 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

