Functional Microbial Features Driving Community Assembly During Seed Germination and Emergence
- PMID: 30008730
- PMCID: PMC6034153
- DOI: 10.3389/fpls.2018.00902
Functional Microbial Features Driving Community Assembly During Seed Germination and Emergence
Abstract
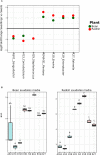
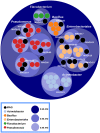
Microbial interactions occurring on and around seeds are especially important for plant fitness since seed-borne microorganisms are the initial source of inoculum for the plant microbiota. In this study, we analyze structural and functional changes occurring within the plant microbiota at these early stages of the plant cycle, namely germination and emergence. To this purpose, we performed shotgun DNA sequencing of microbial assemblages associated to seeds, germinating seeds and seedlings of two plant species: bean and radish. We observed an enrichment of Enterobacteriales and Pseudomonadales during emergence and a set of functional traits linked to copiotrophy that could be responsible for this selection as a result of an increase of nutrient availability after germination. Representative bacterial isolates of taxa that are selected in seedlings showed indeed faster bacterial growth rate in comparison to seed-associated bacteria isolates. Finally, binning of metagenomics contigs results in the reconstruction of population genomes of the major bacterial taxa associated to the samples. Together, our results demonstrate that, although seed microbiota varied across plant species, nutrient availability during germination elicits changes of the composition of microbial communities by potentially selecting microbial groups with functional traits linked to copiotrophy. The data presented here represents the first attempts to empirically assess changes in the microbial community during plant emergence and moves us toward a more holistic understanding of the plant microbiome.
Keywords: copiotrophy; metagenomics; plant microbial communities; seed germination; seed microbiome.
Figures



References
-
- Adam E., Bernhart M., Müller H., Winkler J., Berg G. (2016). The Cucurbita pepo seed microbiome: genotype-specific composition and implications for breeding. Plant Soil 422 1–15.
-
- Adeolu M., Alnajar S., Naushad S., Gupta R. (2016). Genome based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morgane. Int. J. Syst. Evol. Microbiol. 66 5575–5599. 10.1099/ijsem.0.001485 - DOI - PubMed
-
- Bacilio-Jiménez M., Aguilar-Flores S., del Valle M., Pérez A., Zepeda A., Zenteno E. (2001). Endophytic bacteria in rice seeds inhibit early colonization of roots by Azospirillum brasilense. Soil Biol. Biochem 33 167–172. 10.1016/S0038-0717(00)00126-7 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

