A meta-analysis of transcriptome datasets characterizes malignant transformation from melanocytes and nevi to melanoma
- PMID: 30008882
- PMCID: PMC6036496
- DOI: 10.3892/ol.2018.8861
A meta-analysis of transcriptome datasets characterizes malignant transformation from melanocytes and nevi to melanoma
Abstract
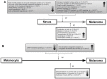
Melanoma represents one of the most aggressive malignancies and has a high tendency to metastasize. The present study aims to investigate the molecular mechanisms of two pathways to cancer transformation with the purpose of identifying potential biomarkers. Our approach is based on a meta-analysis of gene expression profiling contrasting two scenarios: A model that describes a transformation pathway from melanocyte to melanoma and a second model where transformation occurs through an intermediary nevus. Data consists of three independent, publicly available microarray datasets from the Gene Expression Omnibus (GEO) database comprising samples from melanocytes, nevi and melanoma. The present analysis identified 808 differentially expressed genes (528 upregulated and 360 downregulated) in melanoma compared with nevi, and 2,331 differentially expressed genes (946 upregulated and 1,385 downregulated) in melanoma compared with melanocytes. Further analysis narrowed down this list, since 682 differentially expressed genes were found in both models (417 upregulated and 265 downregulated). Enrichment analysis identified relevant dysregulated pathways. This article also presented a discussion on significant genes including ADAM like decysin 1, neudesin neurotrophic factor, MMP19, apolipoprotein L6, C-X-C motif chemokine ligand (CXCL)8, basic, immunoglobulin-like variable motif containing and CXCL16. These are of particular interest because they encode secreted proteins hence represent potential blood biomarkers for the early detection of malignant transformation in both scenarios. Cytotoxic T-lymphocyte associated protein 4, an important therapeutic target in melanoma treatment, was also upregulated in both comparisons indicating a potential involvement in immune tolerance, not only at advanced stages but also during the early transformation to melanoma. The results of the present study may provide a research direction for studying the mechanisms underlying the development of melanoma, depending on its origin.
Keywords: biomarker; melanocyte; melanoma; meta-analysis; nevus; transcriptome.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
