A novel mammalian cell line development platform utilizing nanofluidics and optoelectro positioning technology
- PMID: 30009534
- PMCID: PMC6585769
- DOI: 10.1002/btpr.2690
A novel mammalian cell line development platform utilizing nanofluidics and optoelectro positioning technology
Abstract
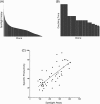
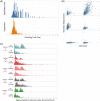
Generating a highly productive cell line is resource intensive and typically involves long timelines because of the need to screen large numbers of candidates in protein production studies. This has led to miniaturization and automation strategies to allow for reductions in resources and higher throughput. Current approaches rely on the use of standard cell culture vessels and bulky liquid handling equipment. New nanofludic technologies offer novel solutions to surpass these limits, further miniaturizing cell culture volumes (105 times smaller) by growing cells on custom nanofluidic chips. Berkeley Lights' OptoElectro Positioning technology projects light patterns to activate photoconductors that gently repel cells to manipulate single cells on nanofluidic culturing chips. Using a fully integrated technology platform (Beacon), common cell culture tasks can be programmed through software, allowing maintenance and analysis of thousands of cell lines in parallel on a single chip. Here, we describe the ability to perform key cell line development work on the Beacon platform. We demonstrate that commercial production Chinese hamster ovary cell lines can be isolated, cultured, screened, and exported at high efficiency. We compare this process head to head with a FACS-enabled microtiter plate-based workflow and demonstrate generation of comparable clonal cell lines with reduced resources. © 2018 American Institute of Chemical Engineers Biotechnol. Prog., 34:1438-1446, 2018.
Keywords: Cell line development; automation; nanofluidics.
© 2018 The Authors. Biotechnology Progress published by Wiley Periodicals, Inc. on behalf of American Institute of Chemical Engineers.
Figures



References
-
- Wurm FM. Production of recombinant protein therapeutics in cultivated mammalian cells. Nat Biotechnol. 2004;22(11):1393–1398. - PubMed
-
- Frye C, Deshpande R, Estes S, Francissen K, Joly J, Lubiniecki A, Munro T, Russell R, Wang T, Anderson K. Industry view on the relative importance of "clonality" of biopharmaceutical‐producing cell lines. Biologicals. 2016;44(2):117–122. - PubMed
-
- Zhou Y, Shaw D, Lam C, Tsukuda J, Yim M, Tang D, Louie S, Laird MW, Snedecor B, Misaghi S. Beating the odds: the poisson distribution of all input cells during limiting dilution grossly underestimates whether a cell line is clonally‐derived or not. Biotechnol Progress. 2017. - PubMed
-
- DeMaria CT, Cairns V, Schwarz C, Zhang J, Guerin M, Zuena E, Estes S, Karey KP. Accelerated clone selection for recombinant CHO CELLS using a FACS‐based high‐throughput screen. Biotechnol Progress. 2007;23(2):465–472. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

