The Role of Functional Amyloids in Bacterial Virulence
- PMID: 30009771
- PMCID: PMC6173799
- DOI: 10.1016/j.jmb.2018.07.010
The Role of Functional Amyloids in Bacterial Virulence
Abstract
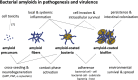
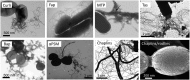
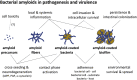
Amyloid fibrils are best known as a product of human and animal protein misfolding disorders, where amyloid formation is associated with cytotoxicity and disease. It is now evident that for some proteins, the amyloid state constitutes the native structure and serves a functional role. These functional amyloids are proving widespread in bacteria and fungi, fulfilling diverse functions as structural components in biofilms or spore coats, as toxins and surface-active fibers, as epigenetic material, peptide reservoirs or adhesins mediating binding to and internalization into host cells. In this review, we will focus on the role of functional amyloids in bacterial pathogenesis. The role of functional amyloids as virulence factor is diverse but mostly indirect. Nevertheless, functional amyloid pathways deserve consideration for the acute and long-term effects of the infectious disease process and may form valid antimicrobial targets.
Keywords: amyloid; bacterial adhesion; bacterial pathogenesis; biofilm; curli.
Copyright © 2018 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures


References
-
- Sipe J.D., Cohen A.S. Review: history of the amyloid fibril. J. Struct. Biol. 2000;130:88–98. - PubMed
-
- Eisenberg D.S., Sawaya M.R. Structural studies of amyloid proteins at the molecular level. Annu. Rev. Biochem. 2017;86:69–95. - PubMed
-
- Chiti F., Dobson C.M. Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu. Rev. Biochem. 2017;86:27–68. - PubMed
-
- Fowler D.M., Koulov A.V., Balch W.E., Kelly J.W. Functional amyloid—from bacteria to humans. Trends Biochem. Sci. 2007;32:217–224. - PubMed
-
- Larsen P., Nielsen J.L., Dueholm M.S., Wetzel R., Otzen D., Nielsen P.H. Amyloid adhesins are abundant in natural biofilms. Environ. Microbiol. 2007;9:3077–3090. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

