Metagenomic binning through low-density hashing
- PMID: 30010790
- PMCID: PMC6330020
- DOI: 10.1093/bioinformatics/bty611
Metagenomic binning through low-density hashing
Abstract
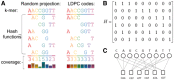
Motivation: Vastly greater quantities of microbial genome data are being generated where environmental samples mix together the DNA from many different species. Here, we present Opal for metagenomic binning, the task of identifying the origin species of DNA sequencing reads. We introduce 'low-density' locality sensitive hashing to bioinformatics, with the addition of Gallager codes for even coverage, enabling quick and accurate metagenomic binning.
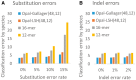
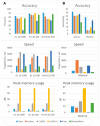
Results: On public benchmarks, Opal halves the error on precision/recall (F1-score) as compared with both alignment-based and alignment-free methods for species classification. We demonstrate even more marked improvement at higher taxonomic levels, allowing for the discovery of novel lineages. Furthermore, the innovation of low-density, even-coverage hashing should itself prove an essential methodological advance as it enables the application of machine learning to other bioinformatic challenges.
Availability and implementation: Full source code and datasets are available at http://opal.csail.mit.edu and https://github.com/yunwilliamyu/opal.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures
References
-
- Altschul S.F., et al. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. - PubMed
-
- Alneberg J., et al. (2014) Binning metagenomic contigs by coverage and composition. Nat. Methods, 11, 1144. - PubMed
-
- Andoni A., Indyk P. (2006) Near-optimal hashing algorithms for approximate nearest neighbor in high dimension. In Foundations of Computer Science, FOCS'06. 47th Annual IEEE Symposium on, pp. 459–468. IEEE.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

