Genome-Wide Sequence Analysis of Kaposi Sarcoma-Associated Herpesvirus Shows Diversification Driven by Recombination
- PMID: 30010810
- PMCID: PMC6195662
- DOI: 10.1093/infdis/jiy427
Genome-Wide Sequence Analysis of Kaposi Sarcoma-Associated Herpesvirus Shows Diversification Driven by Recombination
Abstract
Background: Kaposi sarcoma-associated herpesvirus (KSHV) establishes lifelong infection in the human host and has been associated with a variety of malignancies. KSHV displays striking geographic variation in prevalence, which is highest in sub-Saharan Africa. The current KSHV genome sequences available are all tumor cell line-derived or primary tumor-associated viruses, which have provided valuable insights into KSHV genetic diversity.
Methods: Here, we sequenced 45 KSHV genomes from a Ugandan population cohort in which KSHV is endemic; these are the only genome sequences obtained from nondiseased individuals and of KSHV DNA isolated from saliva.
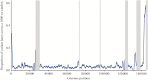
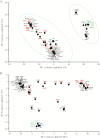
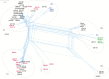
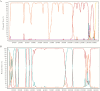
Results: Population structure analysis, along with the 25 published genome sequences from other parts of the world, showed whole-genome variation, separating sequences and variation within the central genome contributing to clustering of genomes by geography. We reveal new evidence for the presence of intragenic recombination and multiple recombination events contributing to the divergence of genomes into at least 5 distinct types.
Discussion: This study shows that large-scale genome-wide sequencing from clinical and epidemiological samples is necessary to capture the full extent of genetic diversity of KSHV, including recombination, and provides evidence to suggest a revision of KSHV genotype nomenclature.
Figures
References
-
- Chang Y, Cesarman E, Pessin MS, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 1994; 266:1865–9. - PubMed
-
- Kaposi M. Idiopathic multiple pigmented sarcoma of the skin. CA Cancer J Clin 1982; 32:342–7. - PubMed
-
- Moore PS, Chang Y. Detection of herpesvirus-like DNA sequences in Kaposi’s sarcoma in patients with and those without HIV infection. N Engl J Med 1995; 332:1181–5. - PubMed
-
- Cesarman E, Mesri EA. Kaposi sarcoma-associated herpesvirus and other viruses in human lymphomagenesis. Curr Top Microbiol Immunol 2007; 312:263–87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

