DI-tector: defective interfering viral genomes' detector for next-generation sequencing data
- PMID: 30012569
- PMCID: PMC6140465
- DOI: 10.1261/rna.066910.118
DI-tector: defective interfering viral genomes' detector for next-generation sequencing data
Abstract
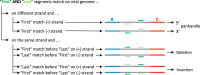
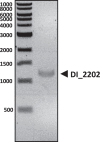
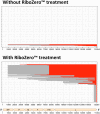
Defective interfering (DI) genomes, or defective viral genomes (DVGs), are truncated viral genomes generated during replication of most viruses, including live viral vaccines. Among these, "panhandle" or copy-back (cb) and "hairpin" or snap-back (sb) DI genomes are generated during RNA virus replication. 5' cb/sb DI genomes are highly relevant for viral pathogenesis since they harbor immunostimulatory properties that increase virus recognition by the innate immune system of the host. We have developed DI-tector, a user-friendly and freely available program that identifies and characterizes cb/sb genomes from next-generation sequencing (NGS) data. DI-tector confirmed the presence of 5' cb genomes in cells infected with measles virus (MV). DI-tector also identified a novel 5' cb genome, as well as a variety of 3' cb/sb genomes whose existence had not previously been detected by conventional approaches in MV-infected cells. The presence of these novel cb/sb genomes was confirmed by RT-qPCR and RT-PCR, validating the ability of DI-tector to reveal the landscape of DI genome population in infected cell samples. Performance assessment using different experimental and simulated data sets revealed the robust specificity and sensitivity of DI-tector. We propose DI-tector as a universal tool for the unbiased detection of DI viral genomes, including 5' cb/sb DI genomes, in NGS data.
Keywords: NGS; copy-back; defective interfering viral genome; panhandle; snap-back; viral replication.
© 2018 Beauclair et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures





References
-
- Auguie B. 2017. gridExtra: miscellaneous functions for “grid” graphics. R package version 2.3. https://CRAN.R-project.org/package=gridExtra.
-
- Bellocq C, Mottet G, Roux L. 1990. Wide occurrence of measles virus subgenomic RNAs in attenuated live-virus vaccines. Biologicals 18: 337–343. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
