Identification of VAPA and VAPB as Kv2 Channel-Interacting Proteins Defining Endoplasmic Reticulum-Plasma Membrane Junctions in Mammalian Brain Neurons
- PMID: 30012696
- PMCID: PMC6113906
- DOI: 10.1523/JNEUROSCI.0893-18.2018
Identification of VAPA and VAPB as Kv2 Channel-Interacting Proteins Defining Endoplasmic Reticulum-Plasma Membrane Junctions in Mammalian Brain Neurons
Abstract
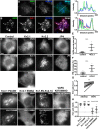
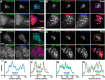
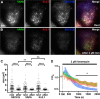
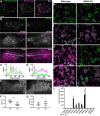
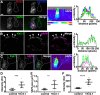
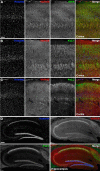
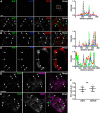
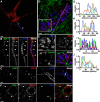
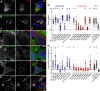
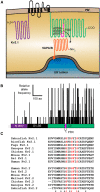
Membrane contacts between endoplasmic reticulum (ER) and plasma membrane (PM), or ER-PM junctions, are ubiquitous in eukaryotic cells and are platforms for lipid and calcium signaling and homeostasis. Recent studies have revealed proteins crucial to the formation and function of ER-PM junctions in non-neuronal cells, but little is known of the ER-PM junctions prominent in aspiny regions of mammalian brain neurons. The Kv2.1 voltage-gated potassium channel is abundantly clustered at ER-PM junctions in brain neurons and is the first PM protein that functions to organize ER-PM junctions. However, the molecular mechanism whereby Kv2.1 localizes to and remodels these junctions is unknown. We used affinity immunopurification and mass spectrometry-based proteomics on brain samples from male and female WT and Kv2.1 KO mice and identified the resident ER vesicle-associated membrane protein-associated proteins isoforms A and B (VAPA and VAPB) as prominent Kv2.1-associated proteins. Coexpression with Kv2.1 or its paralog Kv2.2 was sufficient to recruit VAPs to ER-PM junctions. Multiplex immunolabeling revealed colocalization of Kv2.1 and Kv2.2 with endogenous VAPs at ER-PM junctions in brain neurons from male and female mice in situ and in cultured rat hippocampal neurons, and KO of VAPA in mammalian cells reduces Kv2.1 clustering. The association of VAPA with Kv2.1 relies on a "two phenylalanines in an acidic tract" (FFAT) binding domain on VAPA and a noncanonical phosphorylation-dependent FFAT motif comprising the Kv2-specific clustering or PRC motif. These results suggest that Kv2.1 localizes to and organizes neuronal ER-PM junctions through an interaction with VAPs.SIGNIFICANCE STATEMENT Our study identified the endoplasmic reticulum (ER) proteins vesicle-associated membrane protein-associated proteins isoforms A and B (VAPA and VAPB) as proteins copurifying with the plasma membrane (PM) Kv2.1 ion channel. We found that expression of Kv2.1 recruits VAPs to ER-PM junctions, specialized membrane contact sites crucial to distinct aspects of cell function. We found endogenous VAPs at Kv2.1-mediated ER-PM junctions in brain neurons and other mammalian cells and that knocking out VAPA expression disrupts Kv2.1 clustering. We identified domains of VAPs and Kv2.1 necessary and sufficient for their association at ER-PM junctions. Our study suggests that Kv2.1 expression in the PM can affect ER-PM junctions via its phosphorylation-dependent association to ER-localized VAPA and VAPB.
Keywords: ion channel; membrane contact sites; neuron; subcellular.
Copyright © 2018 the authors 0270-6474/18/387562-23$15.00/0.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
