An enhanced CRISPR repressor for targeted mammalian gene regulation
- PMID: 30013045
- PMCID: PMC6129399
- DOI: 10.1038/s41592-018-0048-5
An enhanced CRISPR repressor for targeted mammalian gene regulation
Abstract
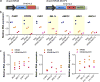
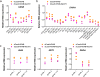
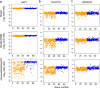
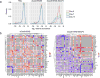
The RNA-guided endonuclease Cas9 can be converted into a programmable transcriptional repressor, but inefficiencies in target-gene silencing have limited its utility. Here we describe an improved Cas9 repressor based on the C-terminal fusion of a rationally designed bipartite repressor domain, KRAB-MeCP2, to nuclease-dead Cas9. We demonstrate the system's superiority in silencing coding and noncoding genes, simultaneously repressing a series of target genes, improving the results of single and dual guide RNA library screens, and enabling new architectures of synthetic genetic circuits.
Conflict of interest statement
G.M.C. is a founder and advisor for Editas Medicine. G.M.C. has equity in Editas and Caribou Biosciences (for full disclosure list, please see:
Figures


References
-
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

