A Novel Multi-Gene Detection Platform for the Analysis of miRNA Expression
- PMID: 30013095
- PMCID: PMC6048151
- DOI: 10.1038/s41598-018-29146-7
A Novel Multi-Gene Detection Platform for the Analysis of miRNA Expression
Abstract
The study of miRNAs and their roles as non-invasive biomarkers has been intensely conducted in cancer diseases over the past decade. Various platforms, ranging from conventional qPCRs to Next Generation Sequencers (NGS), have been widely used to analyze miRNA expression. Here we introduced a novel platform, PanelChip™ Analysis System, which provides a sensitive solution for the analysis of miRNA levels in blood. After conducting miRQC analysis, the system's analytical performance compared favorably against similar nanoscale qPCR-based array technologies. Because PanelChip™ requires only a minimal amount of miRNA for analysis, we used it to screen for potential diagnostic biomarkers in the plasma of patients with oral cavity squamous cell carcinoma (OSCC). Combining the platform with a machine learning algorithm, we were able to discover miRNA expression patterns capable of separating healthy subjects from patients with OSCC.
Conflict of interest statement
The authors declare no competing interests.
Figures
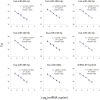

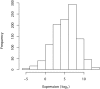
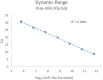
References
Publication types
MeSH terms
Substances
Grants and funding
- CMRPG3E1633, CMRPG3E0512, and CMRPG3G0591-2/Chang Gung Memorial Hospital, Linkou (Linkou Chang Gung Memorial Hospital)/International
- CMRPG3D1063, CMRPG3D1073/Chang Gung Memorial Hospital, Linkou (Linkou Chang Gung Memorial Hospital)/International
- 104-2314-B-182A-073-MY3/Ministry of Science and Technology, Taiwan (Ministry of Science and Technology of Taiwan)/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

