Genome doubling shapes the evolution and prognosis of advanced cancers
- PMID: 30013179
- PMCID: PMC6072608
- DOI: 10.1038/s41588-018-0165-1
Genome doubling shapes the evolution and prognosis of advanced cancers
Abstract
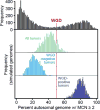
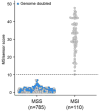
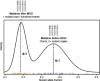
Ploidy abnormalities are a hallmark of cancer, but their impact on the evolution and outcomes of cancers is unknown. Here, we identified whole-genome doubling (WGD) in the tumors of nearly 30% of 9,692 prospectively sequenced advanced cancer patients. WGD varied by tumor lineage and molecular subtype, and arose early in carcinogenesis after an antecedent transforming driver mutation. While associated with TP53 mutations, 46% of all WGD arose in TP53-wild-type tumors and in such cases was associated with an E2F-mediated G1 arrest defect, although neither aberration was obligate in WGD tumors. The variability of WGD across cancer types can be explained in part by cancer cell proliferation rates. WGD predicted for increased morbidity across cancer types, including KRAS-mutant colorectal cancers and estrogen receptor-positive breast cancers, independently of established clinical prognostic factors. We conclude that WGD is highly common in cancer and is a macro-evolutionary event associated with poor prognosis across cancer types.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


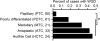






References
-
- Van de Peer Y, Mizrachi E, Marchal K. The evolutionary significance of polyploidy. Nat Rev Genet. 2017;18:411–424. - PubMed
-
- Davoli T, de Lange T. The causes and consequences of polyploidy in normal development and cancer. Annu Rev Cell Dev Biol. 2011;27:585–610. - PubMed
-
- Storchova Z, Pellman D. From polyploidy to aneuploidy, genome instability and cancer. Nat Rev Mol Cell Biol. 2004;5:45–54. - PubMed
-
- Fujiwara T, et al. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

