Genome-wide association and HLA fine-mapping studies identify risk loci and genetic pathways underlying allergic rhinitis
- PMID: 30013184
- PMCID: PMC7068780
- DOI: 10.1038/s41588-018-0157-1
Genome-wide association and HLA fine-mapping studies identify risk loci and genetic pathways underlying allergic rhinitis
Erratum in
-
Author Correction: Genome-wide association and HLA fine-mapping studies identify risk loci and genetic pathways underlying allergic rhinitis.Nat Genet. 2018 Sep;50(9):1343. doi: 10.1038/s41588-018-0197-6. Nat Genet. 2018. PMID: 30116036
Abstract
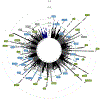
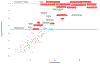
Allergic rhinitis is the most common clinical presentation of allergy, affecting 400 million people worldwide, with increasing incidence in westernized countries1,2. To elucidate the genetic architecture and understand the underlying disease mechanisms, we carried out a meta-analysis of allergic rhinitis in 59,762 cases and 152,358 controls of European ancestry and identified a total of 41 risk loci for allergic rhinitis, including 20 loci not previously associated with allergic rhinitis, which were confirmed in a replication phase of 60,720 cases and 618,527 controls. Functional annotation implicated genes involved in various immune pathways, and fine mapping of the HLA region suggested amino acid variants important for antigen binding. We further performed genome-wide association study (GWAS) analyses of allergic sensitization against inhalant allergens and nonallergic rhinitis, which suggested shared genetic mechanisms across rhinitis-related traits. Future studies of the identified loci and genes might identify novel targets for treatment and prevention of allergic rhinitis.
Conflict of interest statement
Competing financial interests
G.S., I.J., and K.S. are affiliated with deCODE genetics/Amgen declare competing financial interests as employees. C.T., D.A.H., J.Y.T., and the 23andMe Research Team are employees of and hold stock and/or stock options in 23andMe, Inc. L.P. has received a fee for participating in a scientific input engagement meeting from Merck Sharp & Dohme Limited, outside of the submitted work.
Figures
References
-
- Greiner AN, Hellings PW, Rotiroti G & Scadding GK Allergic rhinitis. Lancet 378, 2112–2122 (2011). - PubMed
-
- Björkstén B et al. Worldwide time trends for symptoms of rhinitis and conjunctivitis: Phase III of the International Study of Asthma and Allergies in Childhood. Pediatr. Allergy Immunol. 19, 110–124 (2008). - PubMed
-
- Willemsen G, van Beijsterveldt TCEM, van Baal CGCM, Postma D & Boomsma DI Heritability of self-reported asthma and allergy: a study in adult Dutch twins, siblings and parents. Twin Res. Hum. Genet. 11, 132–142 (2008). - PubMed
-
- Fagnani C et al. Heritability and shared genetic effects of asthma and hay fever: an Italian study of young twins. Twin Res. Hum. Genet 11, 121–131 (2008). - PubMed
-
- Ramasamy A et al. A genome-wide meta-analysis of genetic variants associated with allergic rhinitis and grass sensitization and their interaction with birth order. J. Allergy Clin. Immunol 128, 996–1005 (2011). - PubMed
Methods section references
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

