Host-linked soil viral ecology along a permafrost thaw gradient
- PMID: 30013236
- PMCID: PMC6786970
- DOI: 10.1038/s41564-018-0190-y
Host-linked soil viral ecology along a permafrost thaw gradient
Abstract
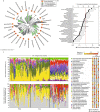
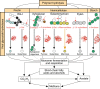
Climate change threatens to release abundant carbon that is sequestered at high latitudes, but the constraints on microbial metabolisms that mediate the release of methane and carbon dioxide are poorly understood1-7. The role of viruses, which are known to affect microbial dynamics, metabolism and biogeochemistry in the oceans8-10, remains largely unexplored in soil. Here, we aimed to investigate how viruses influence microbial ecology and carbon metabolism in peatland soils along a permafrost thaw gradient in Sweden. We recovered 1,907 viral populations (genomes and large genome fragments) from 197 bulk soil and size-fractionated metagenomes, 58% of which were detected in metatranscriptomes and presumed to be active. In silico predictions linked 35% of the viruses to microbial host populations, highlighting likely viral predators of key carbon-cycling microorganisms, including methanogens and methanotrophs. Lineage-specific virus/host ratios varied, suggesting that viral infection dynamics may differentially impact microbial responses to a changing climate. Virus-encoded glycoside hydrolases, including an endomannanase with confirmed functional activity, indicated that viruses influence complex carbon degradation and that viral abundances were significant predictors of methane dynamics. These findings suggest that viruses may impact ecosystem function in climate-critical, terrestrial habitats and identify multiple potential viral contributions to soil carbon cycling.
Conflict of interest statement
The authors declare no competing interests.
Figures


Comment in
-
Permafrost thawing and carbon metabolism.Nat Rev Microbiol. 2018 Sep;16(9):519. doi: 10.1038/s41579-018-0066-4. Nat Rev Microbiol. 2018. PMID: 30042479 No abstract available.
References
-
- Johansson T, et al. Decadal vegetation changes in a northern peatland, greenhouse gas fluxes and net radiative forcing. Glob. Change Biol. 2006;12:2352–2369.
-
- Schuur EAG, et al. Climate change and the permafrost carbon feedback. Nature. 2015;520:171–179. - PubMed
-
- Hultman J, et al. Multi-omics of permafrost, active layer and thermokarst bog soil microbiomes. Nature. 2015;521:208–212. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

