SAV4189, a MarR-Family Regulator in Streptomyces avermitilis, Activates Avermectin Biosynthesis
- PMID: 30013524
- PMCID: PMC6036246
- DOI: 10.3389/fmicb.2018.01358
SAV4189, a MarR-Family Regulator in Streptomyces avermitilis, Activates Avermectin Biosynthesis
Abstract
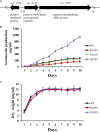
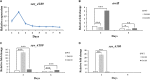
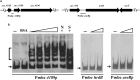
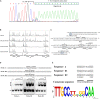
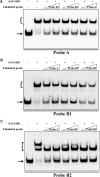
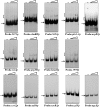
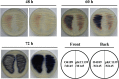
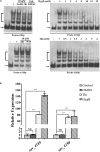
The bacterial species Streptomyces avermitilis is an important industrial producer of avermectins, which are widely utilized as effective anthelmintic and insecticidal drugs. We used gene deletion, complementation, and overexpression experiments to identify SAV4189, a MarR-family transcriptional regulator (MFR) in this species, as an activator of avermectin biosynthesis. SAV4189 indirectly stimulated avermectin production by altering expression of cluster-situated activator gene aveR, and directly repressed the transcription of its own gene (sav_4189) and adjacent cotranscribed gene sav_4190 (which encodes an unknown transmembrane efflux protein). A consensus 13-bp palindromic sequence, 5'-TTGCCYKHRSCAA-3' (Y = T/C; K = T/G; H = A/C/T; R = A/G; S = C/G), was found within the SAV4189-binding sites of its own promoter region, and shown to be essential for binding. The SAV4189 regulon was thus predicted based on bioinformatic analysis. Night new identified SAV4189 targets are involved in transcriptional regulation, primary metabolism, secondary metabolism, and stress response, reflecting a pleiotropic role of SAV4189. sav_4190, the important target gene of SAV4189, exerted a negative effect on avermectin production. sav_4189 overexpression and sav_4190 deletion in S. avermitilis wild-type and industrial strains significantly increased avermectin production. SAV4189 homologs are widespread in other Streptomyces species. sav_4189 overexpression in the model species S. coelicolor also enhanced antibiotic production. The strategy of increasing yield of important antibiotics by engineering of SAV4189 homologs and target gene may potentially be extended to other industrial Streptomyces species. In addition, SAV4189 bound and responded to exogenous antibiotics hygromycin B and thiostrepton to modulate its DNA-binding activity and transcription of target genes. SAV4189 is the first reported exogenous antibiotic receptor among Streptomyces MFRs.
Keywords: MarR-family regulator; SAV4189; Streptomyces avermitilis; avermectins; sav_4190.
Figures

References
-
- Bursy J., Kuhlmann A. U., Pittelkow M., Hartmann H., Jebbar M., Pierik A. J., et al. (2008). Synthesis and uptake of the compatible solutes ectoine and 5-hydroxyectoine by Streptomyces coelicolor A3(2) in response to salt and heat stresses. Appl. Environ. Microbiol. 74 286–7296. 10.1128/AEM.00768-08 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

