Novel Enterobacter Lineage as Leading Cause of Nosocomial Outbreak Involving Carbapenemase-Producing Strains
- PMID: 30014838
- PMCID: PMC6056098
- DOI: 10.3201/eid2408.180151
Novel Enterobacter Lineage as Leading Cause of Nosocomial Outbreak Involving Carbapenemase-Producing Strains
Abstract
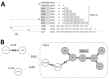
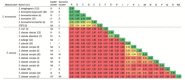
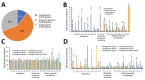
We investigated unusual carbapenemase-producing Enterobacter cloacae complex isolates (n = 8) in the novel sequence type (ST) 873, which caused nosocomial infections in 2 hospitals in France. Whole-genome sequence typing showed the 1-year persistence of the epidemic strain, which harbored a blaVIM-4 ST1-IncHI2 plasmid, in 1 health institution and 2 closely related strains harboring blaCTX-M-15 in the other. These isolates formed a new subgroup in the E. hormaechei metacluster, according to their hsp60 sequences and phylogenomic analysis. The average nucleotide identities, specific biochemical properties, and pangenomic and functional investigations of isolates suggested isolates of a novel species that had acquired genes associated with adhesion and mobility. The emergence of this novel Enterobacter phylogenetic lineage within hospitals should be closely monitored because of its ability to persist and spread.
Keywords: CPE; Carbapenemase; Enterobacter cloacae complex; Enterobacteriaceae; VIM-4; antimicrobial resistance; bacteria; beta-lactamase; carbapenemase-producing Enterobacteriaceae; next-generation sequencing; nosocomial outbreak.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

