Synthetic lethality of TNK2 inhibition in PTPN11-mutant leukemia
- PMID: 30018082
- PMCID: PMC6168748
- DOI: 10.1126/scisignal.aao5617
Synthetic lethality of TNK2 inhibition in PTPN11-mutant leukemia
Abstract
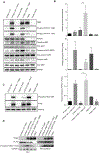
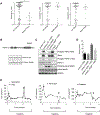
The protein tyrosine phosphatase PTPN11 is implicated in the pathogenesis of juvenile myelomonocytic leukemia (JMML), acute myeloid leukemia (AML), and other malignancies. Activating mutations in PTPN11 increase downstream proliferative signaling and cell survival. We investigated the signaling upstream of PTPN11 in JMML and AML cells and found that PTPN11 was activated by the nonreceptor tyrosine/serine/threonine kinase TNK2 and that PTPN11-mutant JMML and AML cells were sensitive to TNK2 inhibition. In cultured human cell-based assays, PTPN11 and TNK2 interacted directly, enabling TNK2 to phosphorylate PTPN11, which subsequently dephosphorylated TNK2 in a negative feedback loop. Mutations in PTPN11 did not affect this physical interaction but increased the basal activity of PTPN11 such that TNK2-mediated activation was additive. Consequently, coexpression of TNK2 and mutant PTPN11 synergistically increased mitogen-activated protein kinase (MAPK) signaling and enhanced colony formation in bone marrow cells from mice. Chemical inhibition of TNK2 blocked MAPK signaling and colony formation in vitro and decreased disease burden in a patient with PTPN11-mutant JMML who was treated with the multikinase (including TNK2) inhibitor dasatinib. Together, these data suggest that TNK2 is a promising therapeutic target for PTPN11-mutant leukemias.
Copyright © 2018 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures
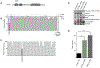

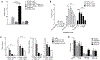
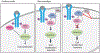
References
-
- Chan G, Kalaitzidis D, Neel BG, The tyrosine phosphatase Shp2 (PTPN11) in cancer. Cancer Metastasis Rev. 27, 179–192 (2008). - PubMed
-
- Kratz CP, Niemeyer CM, Castleberry RP, Cetin M, Bergstrasser E, Emanuel PD, Hasle H, Kardos G, Klein C, Kojima S, Stary J, Trebo M, Zecca M, Gelb BD, Tartaglia M, Loh ML, The mutational spectrum of PTPN11 in juvenile myelomonocytic leukemia and Noonan syndrome/myeloproliferative disease. Blood 106, 2183–2185 (2005). - PMC - PubMed
-
- Stieglitz E, Taylor-Weiner AN, Chang TY, Gelston LC, Wang Y-D, Mazor T, Esquivel E, Yu A, Seepo S, Olsen SR, Rosenberg M, Archambeault SL, Abusin G, Beckman K, Brown PA, Briones M, Carcamo B, Cooper T, Dahl GV, Emanuel PD, Fluchel MN, Goyal RK, Hayashi RJ, Hitzler J, Hugge C, Liu YL, Messinger YH, Mahoney H Jr., Monteleone P, Nemecek ER, Roehrs PA, Schore RJ, Stine KC, Takemoto M, Toretsky JA, Costello JF, Olshen AB, Stewart C, Li Y, Ma J, Gerbing RB, Alonzo TA, Getz G, Gruber TA, Golub TR, Stegmaier K, Loh ML, The genomic landscape of juvenile myelomonocytic leukemia. Nat. Genet. 47, 1326–1333 (2015). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

