PAR-3 controls endothelial planar polarity and vascular inflammation under laminar flow
- PMID: 30018153
- PMCID: PMC6123654
- DOI: 10.15252/embr.201745253
PAR-3 controls endothelial planar polarity and vascular inflammation under laminar flow
Abstract
Impaired cell polarity is a hallmark of diseased tissue. In the cardiovascular system, laminar blood flow induces endothelial planar cell polarity, represented by elongated cell shape and asymmetric distribution of intracellular organelles along the axis of blood flow. Disrupted endothelial planar polarity is considered to be pro-inflammatory, suggesting that the establishment of endothelial polarity elicits an anti-inflammatory response. However, a causative relationship between polarity and inflammatory responses has not been firmly established. Here, we find that a cell polarity protein, PAR-3, is an essential gatekeeper of GSK3β activity in response to laminar blood flow. We show that flow-induced spatial distribution of PAR-3/aPKCλ and aPKCλ/GSK3β complexes controls local GSK3β activity and thereby regulates endothelial planar polarity. The spatial information for GSK3β activation is essential for flow-dependent polarity to the flow axis, but is not necessary for flow-induced anti-inflammatory response. Our results shed light on a novel relationship between endothelial polarity and vascular homeostasis highlighting avenues for novel therapeutic strategies.
Keywords: PAR‐3; atherosclerosis; cell polarity; endothelial cell; flow.
© 2018 The Authors.
Figures
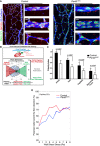
Endothelial axial polarity phenotype in control and Pard3 iΔEC P6 mouse retinal vasculature. Left panels show a large field of view of corresponding retinal vasculature. Right panels show Golgi orientation of ECs in designated areas, corresponding to highlighted areas in left panel (a: artery; v: vein; c: capillary). Green: ERG1/2/3; red: GM130; blue: VE‐cadherin. Arrows are drawn from the center of EC nuclei to the Golgi. Scale bars: 100 μm (left columns, lower magnification) and 20 μm (right columns, higher magnification).
Schema for the Polarity Index calculation. The angle of polarization (α) was calculated from the flow direction and the angle of the nucleus‐to‐Golgi vector.
Analysis of the endothelial Polarity Index, relative to predicted blood flow for each specific vascular bed (n = 3 mice). Data are presented as mean ± SD. P‐values are indicated in the figure (Student's t‐test).
Correlative analysis of wall shear stress and EC polarization in the capillary vascular bed.
Staining of control and Pard3 iΔEC P6 mice retina with adherens junction marker (VE‐cadherin) and endothelial cell marker isolectin‐B4 (iB4).
Staining of control and Pard3 iΔEC P6 retina with basement membrane marker (collagen IV), apical membrane marker (podocalyxin), and isolectin‐B4 (iB4).
Quantification of the number of sprouts/10 μm in angiogenic front. Data are presented as mean ± SEM (n = 3 retinas). Difference **P < 0.01, analyzed by Student's t‐test.
Staining of control and Pard3 iΔEC P6 retina artery with basement membrane marker (collagen IV), apical membrane marker (podocalyxin), adherens junction marker (VE‐cadherin), and nuclei (Hoechst 33342, Sigma). 3D‐reconstituted cross‐section images from the region indicated with white dashed lines are shown in the images.
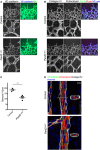
En face confocal microscopy of the aortic arch from the inner curvature to sidewall, stained with anti‐VE‐cadherin (gray), GOLPH4 (red), NF‐κB p65 subunit (green), and ERG1/2/3 (blue) from P56 male mice. Axis of the aorta is indicated on right side of images.
Higher magnification images from (A). Lower panels show p65 signal (gray).
Regional quantification of the cell elongation, cell orientation, Golgi orientation (Polarity Index), and nuclear p65 intensity of ECs.
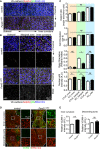
En face confocal microscopy of the inner curvature of the aortic arch and descending aorta from P56 control and Pard3 iΔEC mice. VCAM‐1 is green, and EC nuclei (ERG1/2/3) are red.
Quantification of the relative intensity of VCAM‐1 signal in the ECs of inner curvature of the aorta and descending aorta from control and Pard3 iΔEC mice.

To examine the effect of loss of PAR‐3, the highlighted region of the aorta within the red dotted lines was dissected and analyzed.
To quantify endothelial Golgi orientation toward flow, the angle between the axial polarity vector and the proximal–distal vector (−1 to 1) was analyzed. White arrows indicate the axial polarity vectors from each EC.
Left panel shows the VE‐cadherin signal of aorta ECs. Right panel shows the ellipse fitted to each of the EC, generated by ImageJ software. Red arrows indicate the vectors of the major axis of each ellipse. Distribution of the sin(θ) of the angle between the axial polarity vector and the proximal–distal vector was analyzed (0 to 1).
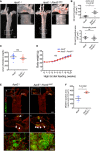
Representative aorta of mice fed with high‐fat diet for 10 weeks (18‐week‐old male mice) stained en face with Oil Red O. Smaller panels show higher magnification images from the aortic arch (1) and descending aorta (2).
Quantification of Oil Red O‐positive regions in aortic arch (upper panel) and descending aorta (lower panel).
Quantification of serum cholesterol level of control (ApoE −/−) and ApoE −/−; Pard3 iΔEC animals after 10 weeks of high‐fat diet feeding.
Body weights of control (ApoE −/−) and ApoE −/−; Pard3 iΔEC animals after 10 weeks of high‐fat diet feeding.
Representative aorta of mice fed with high‐fat diet for 10 weeks (18‐week‐old male mice) stained en face with monocyte/macrophage marker (MOMA‐2, green) and EC marker (EGR, red). White arrowheads indicate MOMA‐2‐positive areas. Smaller panels (a, b, c, d) show higher magnification images from the aortic arch.
Quantification of MOMA‐2‐positive areas in the aortic arch.
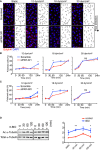
- A
Representative images of flow chamber‐cultured ECs transfected with control (Scrambled) or PARD3‐specific siRNA (siPAR‐3#1) and exposed to the indicated value of shear stress for 60 min. The cells were stained for nuclei (DAPI, blue) and Golgi apparatus (GOLPH4, red). Black arrows indicate corresponding axial polarity vectors. Flow direction is indicated on the right. Scale bar, 35 μm.
- B, C
Axial polarity of ECs treated with control (Scrambled) or PARD3‐specific siRNAs (siPAR‐3#1, B; or siPAR‐3#2, C) in response to 12, 18, and 30 dyn/cm2 laminar flow for the indicated time.
- C
Western blotting of EC lysates treated with 18 dyn/cm2 laminar flow for indicated times, with 1 μM of 6BIO (+) or control (−) containing growth medium. Upper panels show the blot of acetylated α‐tubulin (Ac α‐tubulin), and lower panels show total tubulin. Quantitation of relative intensity of Ac α‐tubulin is shown.

Western blotting of ECs transfected with control scrambled siRNA (indicated as “‐” in the figure) or siPAR‐3#1 and exposed to 18 dyn/cm2 laminar flow.
HEK293 cells were transfected with different amounts of GFP‐PAR‐3 cDNA with HA‐GSK3β and myc‐aPKCλ and were analyzed by Western blotting.
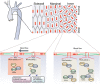
Model for PAR‐3‐mediated GSK3β inactivation. The balance between PAR‐3/aPKCλ complex versus the aPKCλ/GSK3β complex controls GSK3β activity.
HUVECs were transfected with indicated cDNAs and were treated with or without Rho‐kinase inhibitor (Y‐27632, 20 μM). After immunoprecipitation with indicated antibodies, samples were analyzed by Western blotting.
HUVECs were transfected with indicated cDNAs, seeded in the flow chamber, and exposed to 18 dyn/cm2 laminar flow for 60 min. After immunoprecipitation with anti‐myc antibody, samples were analyzed with indicated antibodies by Western blotting. Relative intensity of each signal was statistically analyzed.
RhoA/Rho‐kinase pathway controls GSK3β activity by modulating balance between the PAR‐3/aPKCλ complex versus the aPKCλ/GSK3β complex.
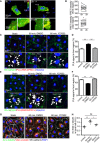
- A
Representative time‐lapse images of ECs transfected with RhoA biosensor under 18 dyn/cm2 laminar flow. FRET/CFP ratio is represented in IMD mode.
- B
Statistical analysis of FRET intensity of EC front and rear region.
- C–F
PLA analysis of Par3 and aPKCλ (C, D) or GSK3β and aPKCλ (E, F) in EC in static and treated with 18 dyn/cm2 laminar flow for 60 min. Arrowheads indicate PLA signal. (D, F) Quantification of the PLA images. Percentile of the PLA signal in the front half of the ECs toward flow was calculated.
- G
Acetylated and total α‐tubulin in ECs under 18 dyn/cm2 flow, with or without Rho‐kinase inhibitor (Y‐27632, 20 μM).
- H
Quantification of regional Ac/total α‐tubulin ratio in the front and rear half of each cells.
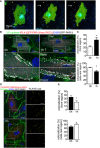
Representative time‐lapse images of ECs transfected with RhoA biosensor. FRET/CFP ratio is represented in IMD mode. Cells were subjected to laminar flow of 12 dyn/cm2 flow for 2 h. Scale bar, 15 μm.
PLA in ECs expressing GFP‐PAR‐3 and myc‐PKCλ in static and under 18 dyn/cm2 flow for 1 h. Arrowheads indicate PAR‐3/aPKCλ PLA signal colocalized with EC junction. Scale bar, 20 μm.
Quantification of the images shown in (B).
Representative images of PLA in ECs expressing GFP‐GSK3β and myc‐PKCλ in static (0 h) and after 1 h subjected to 18 dyn/cm2 flow. EC junction (VE‐cadherin) is green, nuclear stain (DAPI) is blue, and GFP‐GSK3β is gray. Right panels show higher magnification images of the indicated areas. Yellow signals indicate PLA signal colocalized with EC junction. Scale bars, 10 μm (left panel) and 20 μm (right panel).
Quantification of percentile of the PLA signals in the front and rear region of the ECs under static and 1‐h treatment with flow.
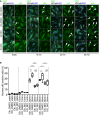
Representative images of control and Pard3KO ECs treated with control (DMSO) or GSK3β inhibitor (6‐BIO, 1 μM) under static conditions or after subjected to 12 dyn/cm2 flow for the indicated times. EC junction (VE‐cadherin) is gray, NF‐κB p65 subunit (p65) is green, and nuclear stain (DAPI) is blue. Scale bar, 50 μm. White arrowheads indicate p65‐positive nuclei.
Quantitative analysis of the percentile of p65‐positive ECs. Data are shown as box and whisker plots. The box spans the interquartile range, horizontal central values are median, and the whiskers extend to the highest and lowest observations (n = 50 cells); statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001) was evaluated by one‐way ANOVA and Tukey's multiple comparison post hoc analysis.
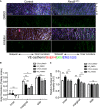
- A
En face confocal microscopy of the aortic arch from the inner curvature to sidewall of control and mutant mice treated with GSK3β inhibitor. Scale bar: 50 μm.
- B, C
Regional quantification of Golgi orientation (Polarity Index, B) and nuclear p65 intensity (C) of ECs. Data are presented as mean ± SEM (n = 5 mice, n = 50 cells from each region). Differences: *P < 0.05, **P < 0.01, analyzed with two‐way ANOVA with Tukey's multiple comparison post hoc analysis.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

