Single cell transcriptome profiling of retinal ganglion cells identifies cellular subtypes
- PMID: 30018341
- PMCID: PMC6050223
- DOI: 10.1038/s41467-018-05134-3
Single cell transcriptome profiling of retinal ganglion cells identifies cellular subtypes
Erratum in
-
Author Correction: Single cell transcriptome profiling of retinal ganglion cells identifies cellular subtypes.Nat Commun. 2018 Aug 7;9(1):3203. doi: 10.1038/s41467-018-05792-3. Nat Commun. 2018. PMID: 30087343 Free PMC article.
Abstract
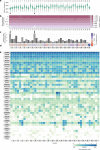
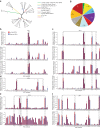
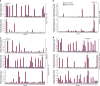
Retinal ganglion cells (RGCs) convey the major output of information collected from the eye to the brain. Thirty subtypes of RGCs have been identified to date. Here, we analyze 6225 RGCs (average of 5000 genes per cell) from right and left eyes by single-cell RNA-seq and classify them into 40 subtypes using clustering algorithms. We identify additional subtypes and markers, as well as transcription factors predicted to cooperate in specifying RGC subtypes. Zic1, a marker of the right eye-enriched subtype, is validated by immunostaining in situ. Runx1 and Fst, the markers of other subtypes, are validated in purified RGCs by fluorescent in situ hybridization (FISH) and immunostaining. We show the extent of gene expression variability needed for subtype segregation, and we show a hierarchy in diversification from a cell-type population to subtypes. Finally, we present a website for comparing the gene expression of RGC subtypes.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

