Proteomic Analysis and Functional Characterization of P4-ATPase Phospholipid Flippases from Murine Tissues
- PMID: 30018401
- PMCID: PMC6050252
- DOI: 10.1038/s41598-018-29108-z
Proteomic Analysis and Functional Characterization of P4-ATPase Phospholipid Flippases from Murine Tissues
Abstract
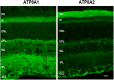
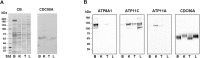
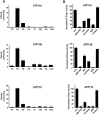
P4-ATPases are a subfamily of P-type ATPases that flip phospholipids across membranes to generate lipid asymmetry, a property vital to many cellular processes. Mutations in several P4-ATPases have been linked to severe neurodegenerative and metabolic disorders. Most P4-ATPases associate with one of three accessory subunit isoforms known as CDC50A (TMEM30A), CDC50B (TMEM30B), and CDC50C (TMEM30C). To identify P4-ATPases that associate with CDC50A, in vivo, and determine their tissue distribution, we isolated P4-ATPases-CDC50A complexes from retina, brain, liver, testes, and kidney on a CDC50A immunoaffinity column and identified and quantified P4-ATPases from their tryptic peptides by mass spectrometry. Of the 12 P4-ATPase that associate with CDC50 subunits, 10 P4-ATPases were detected. Four P4-ATPases (ATP8A1, ATP11A, ATP11B, ATP11C) were present in all five tissues. ATP10D was found in low amounts in liver, brain, testes, and kidney, and ATP8A2 was present in significant amounts in retina, brain, and testes. ATP8B1 was detected only in liver, ATP8B3 and ATP10A only in testes, and ATP8B2 primarily in brain. We also show that ATP11A, ATP11B and ATP11C, like ATP8A1 and ATP8A2, selectively flip phosphatidylserine and phosphatidylethanolamine across membranes. These studies provide new insight into the tissue distribution, relative abundance, subunit interactions and substrate specificity of P4-ATPase-CDC50A complexes.
Conflict of interest statement
The authors declare no competing interests.
Figures


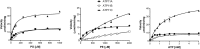
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

