Karyotype evolution in Fusarium
- PMID: 30018869
- PMCID: PMC6048573
- DOI: 10.5598/imafungus.2018.09.01.02
Karyotype evolution in Fusarium
Abstract
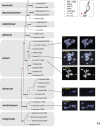
The germ tube burst method (GTBM) was employed to examine karyotypes of 33 Fusarium species representative of 11 species complexes that span the phylogenetic breadth of the genus. The karyotypes revealed that the nucleolar organizing region (NOR), which includes the ribosomal rDNA region, was telomeric in the species where it was discernible. Variable karyotypes were detected in eight species due to variation in numbers of putative core and/or supernumerary chromosomes. The putative core chromosome number (CN) was most variable in the F. solani (CN = 9‒12) and F. buharicum (CN = 9+1 and 18-20) species complexes. Quantitative real-time PCR and genome sequence analysis rejected the hypothesis that the latter variation in CN was due to diploidization. The core CN in six other species complexes where two or more karyotypes were obtained was less variable or fixed. Karyotypes of 10 species in the sambucinum species complex, which is the most derived lineage of Fusarium, revealed that members of this complex possess the lowest CN in the genus. When viewed in context of the species phylogeny, karyotype evolution in Fusarium appears to have been dominated by a reduction in core CN in five closely related complexes that share a most recent common ancestor (tricinctum and incarnatum-equiseti CN = 8-9, chlamydosporum CN = 8, heterosporum CN = 7, sambucinum CN = 4-5) but not in the sister to these complexes (nisikadoi CN = 11, oxysporum CN = 11 and fujikuroi CN = 10-12). CN stability is best illustrated by the F. sambucinum subclade, where the only changes observed since it diverged from other fusaria appear to have involved two independent putative telomere to telomere fusions that reduced the core CN from five to four, once each in the sambucinum and graminearum subclades. Results of the present study indicate a core CN of 4 may be fixed in the latter subclade, which is further distinguished by the absence of putative supernumerary chromosomes. Karyotyping of fusaria in the not too distant future will be done by whole-genome sequencing such that each scaffold represents a complete chromosome from telomere to telomere. The CN data presented here should be of value to assist such full genome assembling.
Keywords: NOR; RPB1; RPB2; accessory; chromosome; genome; pathogen; phylogeny; qPCR; supernumerary.
Figures



References
-
- Akamatsu H, Taga M, Kodama M, Johnson R, Otani H, Kohmoto K. (1999) Molecular karyotypes for Alternaria plant pathogens known to produce host-specific toxins. Current Genetics 35: 647–656. - PubMed
-
- Aoki T, O’Donnell K. (1999) Morphological and molecular characterization of Fusarium pseudograminearum sp. nov., formerly recognized as the group 1 population of F. graminearum. Mycologia 91: 597–609.
-
- Aoki T, O’Donnell K, Geiser DM. (2014) Systematics of key phytopathogenic Fusarium species: current status and future challenges. Journal of General Plant Pathology 80: 189–201.
-
- Boehm EWA, Ploetz RC, Kistler HC. (1994) Statistical analysis of electrophoretic karyotype variation among vegetative compatibility groups of Fusarium oxysporum f. sp. cubense. Molecular Plant-Microbe Interactions 7: 196–207.
LinkOut - more resources
Full Text Sources
Other Literature Sources
