Ustilago species causing leaf-stripe smut revisited
- PMID: 30018872
- PMCID: PMC6048562
- DOI: 10.5598/imafungus.2018.09.01.05
Ustilago species causing leaf-stripe smut revisited
Abstract
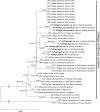
Leaf-stripe smuts on grasses are a highly polyphyletic group within Ustilaginomycotina, occurring in three genera, Tilletia, Urocystis, and Ustilago. Currently more than 12 Ustilago species inciting stripe smuts are recognised. The majority belong to the Ustilago striiformis-complex, with about 30 different taxa described from 165 different plant species. This study aims to assess whether host distinct-lineages can be observed amongst the Ustilago leaf-stripe smuts using nine different loci on a representative set. Phylogenetic reconstructions supported the monophyly of the Ustilago striiformis-complex that causes leaf-stripe and the polyphyly of other leaf-stripe smuts within Ustilago. Furthermore, smut specimens from the same host genus generally clustered together in well-supported clades that often had available species names for these lineages. In addition to already-named lineages, three new lineages were observed, and described as new species on the basis of host specificity and molecular differences: namely Ustilago jagei sp. nov. on Agrostis stolonifera, U. kummeri sp. nov. on Bromus inermis, and U. neocopinata sp. nov. on Dactylis glomerata.
Keywords: DNA-based taxonomy; Ustilaginaceae; host specificity; molecular species discrimination; multigene phylogeny; new taxa; species complex.
Figures





References
-
- Amundsen K, Warnke S. (2012) Agrostis species relationships based on trnL-trnF and atpI-atpH Intergenic Spacer Regions. Hortscience 47: 18–24.
-
- Begerow D, Stoll M, Bauer R. (2006) A phylogenetic hypothesis of Ustilaginomycotina based on multiple gene analyses and morphological data. Mycologia 98: 906–916. - PubMed
-
- Berkeley MJ, Broome CE. (1850) Notices of British fungi. Annals and Magazine of Natural History 5: 455–467.
-
- Catalán P, Torrecilla P, Ángel J, Rodríguez L, Olmstead RG. (2004) Phylogeny of the festucoid grasses of subtribe Loliinae and allies (Poeae, Pooideae) inferred from ITS and trnL–F sequences. Molecular Phylogenetics and Evolution 31: 517–541. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
