Iron-dependent cleavage of ribosomal RNA during oxidative stress in the yeast Saccharomyces cerevisiae
- PMID: 30021840
- PMCID: PMC6139556
- DOI: 10.1074/jbc.RA118.004174
Iron-dependent cleavage of ribosomal RNA during oxidative stress in the yeast Saccharomyces cerevisiae
Abstract
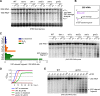
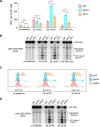
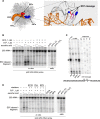
Stress-induced strand breaks in rRNA have been observed in many organisms, but the mechanisms by which they originate are not well-understood. Here we show that a chemical rather than an enzymatic mechanism initiates rRNA cleavages during oxidative stress in yeast (Saccharomyces cerevisiae). We used cells lacking the mitochondrial glutaredoxin Grx5 to demonstrate that oxidant-induced cleavage formation in 25S rRNA correlates with intracellular iron levels. Sequestering free iron by chemical or genetic means decreased the extent of rRNA degradation and relieved the hypersensitivity of grx5Δ cells to the oxidants. Importantly, subjecting purified ribosomes to an in vitro iron/ascorbate reaction precisely recapitulated the 25S rRNA cleavage pattern observed in cells, indicating that redox activity of the ribosome-bound iron is responsible for the strand breaks in the rRNA. In summary, our findings provide evidence that oxidative stress-associated rRNA cleavages can occur through rRNA strand scission by redox-active, ribosome-bound iron that potentially promotes Fenton reaction-induced hydroxyl radical production, implicating intracellular iron as a key determinant of the effects of oxidative stress on ribosomes. We propose that iron binding to specific ribosome elements primes rRNA for cleavages that may play a role in redox-sensitive tuning of the ribosome function in stressed cells.
Keywords: Fenton reaction; RNA; RNA degradation; cell death; glutaredoxin; iron; iron homeostasis; iron metabolism; oxidative stress; reactive oxygen species (ROS); ribosome; yeast.
© 2018 Zinskie et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health
Figures



References
-
- Topf U., Suppanz I., Samluk L., Wrobel L., Böser A., Sakowska P., Knapp B., Pietrzyk M. K., Chacinska A., and Warscheid B. (2018) Quantitative proteomics identifies redox switches for global translation modulation by mitochondrially produced reactive oxygen species. Nat. Commun. 9, 324 10.1038/s41467-017-02694-8 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

