Gene Level Regulation of Na,K-ATPase in the Renal Proximal Tubule Is Controlled by Two Independent but Interacting Regulatory Mechanisms Involving Salt Inducible Kinase 1 and CREB-Regulated Transcriptional Coactivators
- PMID: 30021947
- PMCID: PMC6073390
- DOI: 10.3390/ijms19072086
Gene Level Regulation of Na,K-ATPase in the Renal Proximal Tubule Is Controlled by Two Independent but Interacting Regulatory Mechanisms Involving Salt Inducible Kinase 1 and CREB-Regulated Transcriptional Coactivators
Abstract
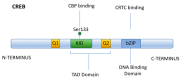
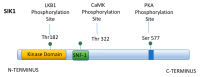
For many years, studies concerning the regulation of Na,K-ATPase were restricted to acute regulatory mechanisms, which affected the phosphorylation of Na,K-ATPase, and thus its retention on the plasma membrane. However, in recent years, this focus has changed. Na,K-ATPase has been established as a signal transducer, which becomes part of a signaling complex as a consequence of ouabain binding. Na,K-ATPase within this signaling complex is localized in caveolae, where Na,K-ATPase has also been observed to regulate Inositol 1,4,5-Trisphosphate Receptor (IP3R)-mediated calcium release. This latter association has been implicated as playing a role in signaling by G Protein Coupled Receptors (GPCRs). Here, the consequences of signaling by renal effectors that act via such GPCRs are reviewed, including their regulatory effects on Na,K-ATPase gene expression in the renal proximal tubule (RPT). Two major types of gene regulation entail signaling by Salt Inducible Kinase 1 (SIK1). On one hand, SIK1 acts so as to block signaling via cAMP Response Element (CRE) Binding Protein (CREB) Regulated Transcriptional Coactivators (CRTCs) and on the other hand, SIK1 acts so as to stimulate signaling via the Myocyte Enhancer Factor 2 (MEF2)/nuclear factor of activated T cell (NFAT) regulated genes. Ultimate consequences of these pathways include regulatory effects which alter the rate of transcription of the Na,K-ATPase β1 subunit gene atp1b1 by CREB, as well as by MEF2/NFAT.
Keywords: CREB; CRTC; Histone Deacetylase; Na,K-ATPase; SIK1; catecholamines; kidney; proximal tubule; renal cell culture.
Conflict of interest statement
The author declares no conflict of interest.
Figures

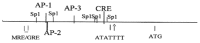



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

