Task-induced brain state manipulation improves prediction of individual traits
- PMID: 30022026
- PMCID: PMC6052101
- DOI: 10.1038/s41467-018-04920-3
Task-induced brain state manipulation improves prediction of individual traits
Abstract
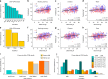
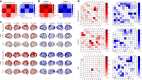
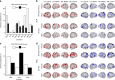
Recent work has begun to relate individual differences in brain functional organization to human behaviors and cognition, but the best brain state to reveal such relationships remains an open question. In two large, independent data sets, we here show that cognitive tasks amplify trait-relevant individual differences in patterns of functional connectivity, such that predictive models built from task fMRI data outperform models built from resting-state fMRI data. Further, certain tasks consistently yield better predictions of fluid intelligence than others, and the task that generates the best-performing models varies by sex. By considering task-induced brain state and sex, the best-performing model explains over 20% of the variance in fluid intelligence scores, as compared to <6% of variance explained by rest-based models. This suggests that identifying and inducing the right brain state in a given group can better reveal brain-behavior relationships, motivating a paradigm shift from rest- to task-based functional connectivity analyses.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Grants and funding
- RC2 MH089924/MH/NIMH NIH HHS/United States
- U54 MH091657/MH/NIMH NIH HHS/United States
- T32 GM007205/GM/NIGMS NIH HHS/United States
- T32GM007205/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)/International
- RC2 MH089983/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

