Pancreatic islets communicate with lymphoid tissues via exocytosis of insulin peptides
- PMID: 30022165
- PMCID: PMC6090537
- DOI: 10.1038/s41586-018-0341-6
Pancreatic islets communicate with lymphoid tissues via exocytosis of insulin peptides
Abstract
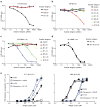
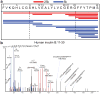
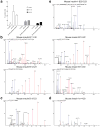
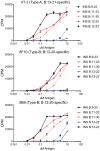
Tissue-specific autoimmunity occurs when selected antigens presented by susceptible alleles of the major histocompatibility complex are recognized by T cells. However, the reason why certain specific self-antigens dominate the response and are indispensable for triggering autoreactivity is unclear. Spontaneous presentation of insulin is essential for initiating autoimmune type 1 diabetes in non-obese diabetic mice1,2. A major set of pathogenic CD4 T cells specifically recognizes the 12-20 segment of the insulin B-chain (B:12-20), an epitope that is generated from direct presentation of insulin peptides by antigen-presenting cells3,4. These T cells do not respond to antigen-presenting cells that have taken up insulin that, after processing, leads to presentation of a different segment representing a one-residue shift, B:13-214. CD4 T cells that recognize B:12-20 escape negative selection in the thymus and cause diabetes, whereas those that recognize B:13-21 have only a minor role in autoimmunity3-5. Although presentation of B:12-20 is evident in the islets3,6, insulin-specific germinal centres can be formed in various lymphoid tissues, suggesting that insulin presentation is widespread7,8. Here we use live imaging to document the distribution of insulin recognition by CD4 T cells throughout various lymph nodes. Furthermore, we identify catabolized insulin peptide fragments containing defined pathogenic epitopes in β-cell granules from mice and humans. Upon glucose challenge, these fragments are released into the circulation and are recognized by CD4 T cells, leading to an activation state that results in transcriptional reprogramming and enhanced diabetogenicity. Therefore, a tissue such as pancreatic islets, by releasing catabolized products, imposes a constant threat to self-tolerance. These findings reveal a self-recognition pathway underlying a primary autoantigen and provide a foundation for assessing antigenic targets that precipitate pathogenic outcomes by systemically sensitizing lymphoid tissues.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





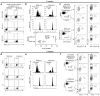
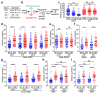



Comment in
-
Peptide secretion triggers diabetes.Nature. 2018 Aug;560(7716):33-34. doi: 10.1038/d41586-018-05710-z. Nature. 2018. PMID: 30061643 No abstract available.
-
Tissue Crosstalk in T1D: Is Insulin Special?Immunity. 2018 Sep 18;49(3):394-396. doi: 10.1016/j.immuni.2018.09.001. Immunity. 2018. PMID: 30231981
References
-
- Unanue ER. Antigen presentation in the autoimmune diabetes of the NOD mouse. Annu Rev Immunol. 2014;32:579–608. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

