The shieldin complex mediates 53BP1-dependent DNA repair
- PMID: 30022168
- PMCID: PMC6141009
- DOI: 10.1038/s41586-018-0340-7
The shieldin complex mediates 53BP1-dependent DNA repair
Abstract
53BP1 is a chromatin-binding protein that regulates the repair of DNA double-strand breaks by suppressing the nucleolytic resection of DNA termini1,2. This function of 53BP1 requires interactions with PTIP3 and RIF14-9, the latter of which recruits REV7 (also known as MAD2L2) to break sites10,11. How 53BP1-pathway proteins shield DNA ends is currently unknown, but there are two models that provide the best potential explanation of their action. In one model the 53BP1 complex strengthens the nucleosomal barrier to end-resection nucleases12,13, and in the other 53BP1 recruits effector proteins with end-protection activity. Here we identify a 53BP1 effector complex, shieldin, that includes C20orf196 (also known as SHLD1), FAM35A (SHLD2), CTC-534A2.2 (SHLD3) and REV7. Shieldin localizes to double-strand-break sites in a 53BP1- and RIF1-dependent manner, and its SHLD2 subunit binds to single-stranded DNA via OB-fold domains that are analogous to those of RPA1 and POT1. Loss of shieldin impairs non-homologous end-joining, leads to defective immunoglobulin class switching and causes hyper-resection. Mutations in genes that encode shieldin subunits also cause resistance to poly(ADP-ribose) polymerase inhibition in BRCA1-deficient cells and tumours, owing to restoration of homologous recombination. Finally, we show that binding of single-stranded DNA by SHLD2 is critical for shieldin function, consistent with a model in which shieldin protects DNA ends to mediate 53BP1-dependent DNA repair.
Conflict of interest statement
DD and TH are advisors to Repare Therapeutics. CJL is a named inventor on patents describing the use of PARP inhibitors (DK3044221 (T3), ES2611504 (T3), US2014378525 (A1), WO2008020180 (A2), WO2009027650 (A1)) and stands to gain from their use as part of the ICR “Rewards to Inventors” scheme.
Figures


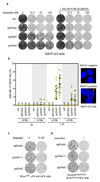
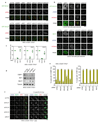

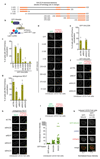





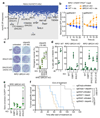

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

