Inhibition of DNA Methylation at the MLH1 Promoter Region Using Pyrrole-Imidazole Polyamide
- PMID: 30023504
- PMCID: PMC6044701
- DOI: 10.1021/acsomega.6b00229
Inhibition of DNA Methylation at the MLH1 Promoter Region Using Pyrrole-Imidazole Polyamide
Abstract
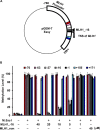
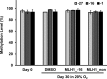
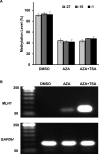
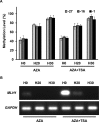
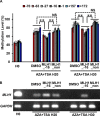
Aberrant DNA methylation causes major epigenetic changes and has been implicated in cancer following the inactivation of tumor suppressor genes by hypermethylation of promoter CpG islands. Although methylated DNA regions can be randomly demethylated by 5-azacytidine and 5-aza-2'-deoxycytidine, site-specific inhibition of DNA methylation, for example, in the promoter region of a specific gene, has yet to be technically achieved. Hairpin pyrrole (Py)-imidazole (Im) polyamides are small molecules that can be designed to recognize and bind to particular DNA sequences. In this study, we synthesized the hairpin polyamide MLH1_-16 (Py-Im-β-Im-Im-Py-γ-Im-Py-β-Im-Py-Py) to target a CpG site 16 bp upstream of the transcription start site of the human MLH1 gene. MLH1 is known to be frequently silenced by promoter hypermethylation, causing microsatellite instability and a hypermutation phenotype in cancer. We show that MLH1_-16 binds to the target site and that CpG methylation around the binding site is selectively inhibited in vitro. MLH1_non, which does not have a recognition site in the MLH1 promoter, neither binds to the sequence nor inhibits DNA methylation in the region. When MLH1_-16 was used to treat RKO human colorectal cancer cells in a remethylating system involving the MLH1 promoter under hypoxic conditions (1% O2), methylation of the MLH1 promoter was inhibited in the region surrounding the compound binding site. Silencing of the MLH1 expression was also inhibited. Promoter methylation and silencing of MLH1 were not inhibited when MLH1_non was added. These results indicate that Py-Im polyamides can act as sequence-specific antagonists of CpG methylation in living cells.
Conflict of interest statement
The authors declare the following competing financial interest(s): As for Inhibition of DNA methylation in living cells, we filed a patent for PIP molecule.
Figures


Similar articles
-
Orientation preferences of hairpin pyrrole-imidazole polyamides toward mCGG site.Bioorg Med Chem. 2019 Jun 1;27(11):2167-2171. doi: 10.1016/j.bmc.2019.04.006. Epub 2019 Apr 10. Bioorg Med Chem. 2019. PMID: 31000407
-
Synthetic pyrrole-imidazole polyamide inhibits expression of the human transforming growth factor-beta1 gene.J Pharmacol Exp Ther. 2005 Nov;315(2):571-5. doi: 10.1124/jpet.105.089086. Epub 2005 Aug 24. J Pharmacol Exp Ther. 2005. PMID: 16120815
-
Development of gene silencing pyrrole-imidazole polyamide targeting the TGF-beta1 promoter for treatment of progressive renal diseases.J Am Soc Nephrol. 2006 Feb;17(2):422-32. doi: 10.1681/ASN.2005060650. Epub 2005 Dec 21. J Am Soc Nephrol. 2006. PMID: 16371433
-
Pyrrole-imidazole hairpin polyamides with high affinity at 5'-CGCG-3' DNA sequence; influence of cytosine methylation on binding.Nucleic Acids Res. 2008 May;36(9):2889-94. doi: 10.1093/nar/gkn116. Epub 2008 Apr 1. Nucleic Acids Res. 2008. PMID: 18385159 Free PMC article.
-
Progress and prospects of pyrrole-imidazole polyamide-fluorophore conjugates as sequence-selective DNA probes.Chembiochem. 2012 Oct 15;13(15):2170-85. doi: 10.1002/cbic.201200451. Epub 2012 Sep 28. Chembiochem. 2012. PMID: 23023993 Review.
Cited by
-
Region-specific alteration of histone modification by LSD1 inhibitor conjugated with pyrrole-imidazole polyamide.Oncotarget. 2018 Jun 29;9(50):29316-29335. doi: 10.18632/oncotarget.25451. eCollection 2018 Jun 29. Oncotarget. 2018. PMID: 30034620 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

