Rad51 recruitment and exclusion of non-homologous end joining during homologous recombination at a Tus/Ter mammalian replication fork barrier
- PMID: 30024881
- PMCID: PMC6067765
- DOI: 10.1371/journal.pgen.1007486
Rad51 recruitment and exclusion of non-homologous end joining during homologous recombination at a Tus/Ter mammalian replication fork barrier
Abstract
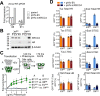
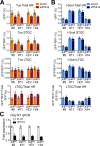
Classical non-homologous end joining (C-NHEJ) and homologous recombination (HR) compete to repair mammalian chromosomal double strand breaks (DSBs). However, C-NHEJ has no impact on HR induced by DNA nicking enzymes. In this case, the replication fork is thought to convert the DNA nick into a one-ended DSB, which lacks a readily available partner for C-NHEJ. Whether C-NHEJ competes with HR at a non-enzymatic mammalian replication fork barrier (RFB) remains unknown. We previously showed that conservative "short tract" gene conversion (STGC) induced by a chromosomal Tus/Ter RFB is a product of bidirectional replication fork stalling. This finding raises the possibility that Tus/Ter-induced STGC proceeds via a two-ended DSB intermediate. If so, Tus/Ter-induced STGC might be subject to competition by C-NHEJ. However, in contrast to the DSB response, where genetic ablation of C-NHEJ stimulates HR, we report here that Tus/Ter-induced HR is unaffected by deletion of either of two C-NHEJ genes, Xrcc4 or Ku70. These results show that Tus/Ter-induced HR does not entail the formation of a two-ended DSB to which C-NHEJ has competitive access. We found no evidence that the alternative end-joining factor, DNA polymerase θ, competes with Tus/Ter-induced HR. We used chromatin-immunoprecipitation to compare Rad51 recruitment to a Tus/Ter RFB and to a neighboring site-specific DSB. Rad51 accumulation at Tus/Ter was more intense and more sustained than at a DSB. In contrast to the DSB response, Rad51 accumulation at Tus/Ter was restricted to within a few hundred base pairs of the RFB. Taken together, these findings suggest that the major DNA structures that bind Rad51 at a Tus/Ter RFB are not conventional DSBs. We propose that Rad51 acts as an "early responder" at stalled forks, binding single stranded daughter strand gaps on the arrested lagging strand, and that Rad51-mediated fork remodeling generates HR intermediates that are incapable of Ku binding and therefore invisible to the C-NHEJ machinery.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

