Molecular and antigenic characterization of group C orthobunyaviruses isolated in Peru
- PMID: 30024910
- PMCID: PMC6053143
- DOI: 10.1371/journal.pone.0200576
Molecular and antigenic characterization of group C orthobunyaviruses isolated in Peru
Abstract
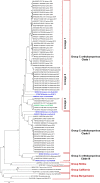
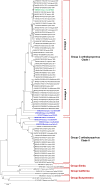
Group C orthobunyaviruses (GRCVs) are a complex of viruses in the genus Orthobunyavirus and are associated with human febrile disease in tropical and subtropical areas of South and Central America. While numerous GRCVs have been isolated from mosquitoes, animals, and humans, genetic analysis of these viruses is limited. In this study, we characterized 65 GRCV isolates from febrile patients identified through clinic-based surveillance in the northern and southern Peruvian Amazon. A 500 base pair region of the S segment and 750 base pair regions of the M and L segments were sequenced. Pairwise sequence analysis of the clinical isolates showed nucleotide identities ranging from 68% to 100% and deduced amino acid sequence identities ranging from 72% to 100%. Sequences were compared with reference strains of the following GRCVs: Caraparu virus (CARV), Murutucu virus (MURV), Oriboca virus (ORIV), Marituba virus (MTBV), Itaqui virus (ITQV), Apeu virus (APEUV), and Madrid virus (MADV). Sequence comparison of clinical isolates with the prototype strains based on the S and L segments identified two clades; clade I included isolates with high genetic association with CARV-MADV, and clade II included isolates with high genetic association with MURV, ORIV, APEUV, and MTBV. Genetic relationships based on the M segment were at time inconsistent with those based on the S and L segments. However, clade groupings based on the M segment were highly consistent with relationships based on microneutralization assays. These results advance our understanding of the genetic and serologic relationships of GRCVs circulating in the Peruvian Amazon.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Casals J, Whitman L. Group C, a new serological group of hitherto undescribed arthropod-borne viruses. Immunological studies. Am J Trop Med Hyg. 1961;10: 250–258. - PubMed
-
- Causey OR, Causey CE, Maroja OM, Macedo DG. The isolation of arthropod-borne viruses, including members of two hitherto undescribed serological groups, in the Amazon region of Brazil. Am J Trop Med Hyg. 1961;10: 227–249. - PubMed
-
- Shope RE, Causey OR. Further studies on the serological relationships of group C arthropod-borne viruses and the application of these relationships to rapid identification of types. Am J Trop Med Hyg. 1962;11: 283–290. - PubMed
-
- Shope RE, Causey CE, Causey OR. Itaqui Virus, a New Member of Arthropod-Borne Group C*. Am J Trop Med Hyg. 1961;10: 264–265.
-
- Shope RE, Woodall JP, Travassos da Rosa A. The epidemiology of diseases caused by viruses in Groups C and Guama (Bunyaviridae) In: Monath TP, editor. The Arboviruses: Epidemiology and Ecology. Boca Raton, Florida, 33431: CRC Press, Inc.; 1988. pp. 37–52.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

