Nuclear-Mitochondrial interactions influence susceptibility to HIV-associated neurocognitive impairment
- PMID: 30026132
- PMCID: PMC6336535
- DOI: 10.1016/j.mito.2018.07.004
Nuclear-Mitochondrial interactions influence susceptibility to HIV-associated neurocognitive impairment
Abstract
HIV-associated neurocognitive impairment (NCI) is a term established to capture a wide spectrum of HIV related neurocognitive deficits ranging in severity from asymptomatic to dementia. The genetic underpinnings of this complex phenotype are incompletely understood. Mitochondrial function has long been thought to play a role in neurodegeneration, along with iron metabolism and transport. In this work, we aimed to characterize the interplay of mitochondrial DNA (mtDNA) haplogroup and nuclear genetic associations to NCI phenotypes in the CHARTER cohort, encompassing 1025 individuals of European-descent, African-descent, or admixed Hispanic. We first employed a polygenic modeling approach to investigate the global effect of previous marginally associated nuclear SNPs, and to examine how the polygenic effect of these SNPs is influenced by mtDNA haplogroups. We see evidence of a significant interaction between nuclear SNPs en masse and mtDNA haplogroups within European-descent and African-descent individuals. Subsequently, we performed an analysis of each SNP by mtDNA haplogroup, and detected significant interactions between two nuclear SNPs (rs17160128 and rs12460243) and European haplogroups. These findings, which require validation in larger cohorts, indicate a potential new role for nuclear-mitochondrial DNA interactions in susceptibility to NCI and shed light onto the pathophysiology of this neurocognitive phenotype.
Keywords: Genomics; HIV; Haplogroups; Neurocognitive impairment; Nuclear mitochondrial interactions; Single nucleotide polymorphism.
Copyright © 2018 The Authors. Published by Elsevier B.V. All rights reserved.
Figures

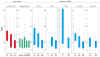
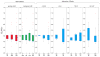

References
-
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

