VHL substrate transcription factor ZHX2 as an oncogenic driver in clear cell renal cell carcinoma
- PMID: 30026228
- PMCID: PMC6154478
- DOI: 10.1126/science.aap8411
VHL substrate transcription factor ZHX2 as an oncogenic driver in clear cell renal cell carcinoma
Abstract
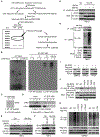
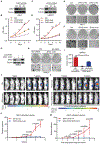
Inactivation of the von Hippel-Lindau (VHL) E3 ubiquitin ligase protein is a hallmark of clear cell renal cell carcinoma (ccRCC). Identifying how pathways affected by VHL loss contribute to ccRCC remains challenging. We used a genome-wide in vitro expression strategy to identify proteins that bind VHL when hydroxylated. Zinc fingers and homeoboxes 2 (ZHX2) was found as a VHL target, and its hydroxylation allowed VHL to regulate its protein stability. Tumor cells from ccRCC patients with VHL loss-of-function mutations usually had increased abundance and nuclear localization of ZHX2. Functionally, depletion of ZHX2 inhibited VHL-deficient ccRCC cell growth in vitro and in vivo. Mechanistically, integrated chromatin immunoprecipitation sequencing and microarray analysis showed that ZHX2 promoted nuclear factor κB activation. These studies reveal ZHX2 as a potential therapeutic target for ccRCC.
Copyright © 2018 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures


Comment in
-
Transcriptional control of kidney cancer.Science. 2018 Jul 20;361(6399):226-227. doi: 10.1126/science.aau4385. Science. 2018. PMID: 30026212 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

