Small molecule activator of Nm23/NDPK as an inhibitor of metastasis
- PMID: 30026594
- PMCID: PMC6053448
- DOI: 10.1038/s41598-018-29101-6
Small molecule activator of Nm23/NDPK as an inhibitor of metastasis
Abstract
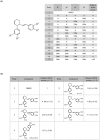
Nm23-H1/NDPK-A is a tumor metastasis suppressor having NDP kinase (NDPK) activity. Nm23-H1 is positively associated with prolonged disease-free survival and good prognosis of cancer patients. Approaches to increasing the cellular levels of Nm23-H1 therefore have significance in the therapy of metastatic cancers. We found a small molecule, (±)-trans-3-(3,4-dimethoxyphenyl)-4-[(E)-3,4-dimethoxystyryl]cyclohex-1-ene, that activates Nm23, hereafter called NMac1. NMac1 directly binds to Nm23-H1 and increases its NDPK activity. Employing various NMac1 derivatives and hydrogen/deuterium mass spectrometry (HDX-MS), we identified the pharmacophore and mode of action of NMac1. We found that NMac1 binds to the C-terminal of Nm23-H1 and induces the NDPK activation through its allosteric conformational changes. NMac1-treated MDA-MB-231 breast cancer cells showed dramatic changes in morphology and actin-cytoskeletal organization following inhibition of Rac1 activation. NMac1 also suppressed invasion and migration in vitro, and metastasis in vivo, in a breast cancer mouse model. NMac1 as an activator of NDPK has potential as an anti-metastatic agent.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Ma D, Luyten GP, Luider TM, Jager MJ, Niederkorn JY. Association between NM23-H1 gene expression and metastasis of human uveal melanoma in an animal model. Invest Ophthalmol Vis Sci. 1996;37(11):2293–2301. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

