All-Optical Assay to Study Biological Neural Networks
- PMID: 30026684
- PMCID: PMC6041400
- DOI: 10.3389/fnins.2018.00451
All-Optical Assay to Study Biological Neural Networks
Abstract
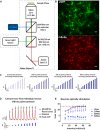
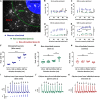
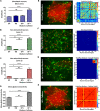
We introduce a novel all-optical assay for functional studies of biological neural networks in vitro. We created a novel optogenetic construct named OptoCaMP which is a combination of a channelrhodopsin variant (CheRiff) and a red genetically encoded calcium indicator (GECI) (jRCaMP1b). It enables simultaneous optical stimulation and recording from large population of neurons with single-cell readout. Additionally, we have developed a spatio-temporal all-optical assay to simultaneously stimulate a sub-section of a neural network and record evoked calcium activity, in both stimulated and non-stimulated neurons, thus allowing the investigation of the spread of excitation through an interconnected network. Finally, we demonstrate the sensitivity of this assay to the change of neural network connectivity.
Keywords: all-optical assay; calcium imaging; connectivity; lentiviral transduction; neural networks; neurons; optogenetics.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

