RSEQREP: RNA-Seq Reports, an open-source cloud-enabled framework for reproducible RNA-Seq data processing, analysis, and result reporting
- PMID: 30026912
- PMCID: PMC6039931
- DOI: 10.12688/f1000research.13049.2
RSEQREP: RNA-Seq Reports, an open-source cloud-enabled framework for reproducible RNA-Seq data processing, analysis, and result reporting
Abstract
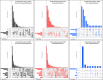
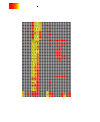
RNA-Seq is increasingly being used to measure human RNA expression on a genome-wide scale. Expression profiles can be interrogated to identify and functionally characterize treatment-responsive genes. Ultimately, such controlled studies promise to reveal insights into molecular mechanisms of treatment effects, identify biomarkers, and realize personalized medicine. RNA-Seq Reports (RSEQREP) is a new open-source cloud-enabled framework that allows users to execute start-to-end gene-level RNA-Seq analysis on a preconfigured RSEQREP Amazon Virtual Machine Image (AMI) hosted by AWS or on their own Ubuntu Linux machine via a Docker container or installation script. The framework works with unstranded, stranded, and paired-end sequence FASTQ files stored locally, on Amazon Simple Storage Service (S3), or at the Sequence Read Archive (SRA). RSEQREP automatically executes a series of customizable steps including reference alignment, CRAM compression, reference alignment QC, data normalization, multivariate data visualization, identification of differentially expressed genes, heatmaps, co-expressed gene clusters, enriched pathways, and a series of custom visualizations. The framework outputs a file collection that includes a dynamically generated PDF report using R, knitr, and LaTeX, as well as publication-ready table and figure files. A user-friendly configuration file handles sample metadata entry, processing, analysis, and reporting options. The configuration supports time series RNA-Seq experimental designs with at least one pre- and one post-treatment sample for each subject, as well as multiple treatment groups and specimen types. All RSEQREP analyses components are built using open-source R code and R/Bioconductor packages allowing for further customization. As a use case, we provide RSEQREP results for a trivalent influenza vaccine (TIV) RNA-Seq study that collected 1 pre-TIV and 10 post-TIV vaccination samples (days 1-10) for 5 subjects and two specimen types (peripheral blood mononuclear cells and B-cells).
Keywords: RNA-Seq; RSEQREP; cloud computing; differential gene expression; pathway enrichment; reproducible research; transcriptomics; trivalent influenza vaccine.
Conflict of interest statement
No competing interests were disclosed.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

