Molecular heterogeneity in human papillomavirus-dependent and -independent vulvar carcinogenesis
- PMID: 30030907
- PMCID: PMC6144162
- DOI: 10.1002/cam4.1633
Molecular heterogeneity in human papillomavirus-dependent and -independent vulvar carcinogenesis
Abstract
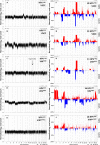
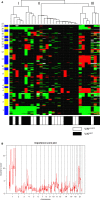
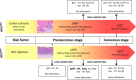
Vulvar squamous cell carcinoma (VSCC) and precancerous vulvar intraepithelial neoplasia (VIN) can develop through human papillomavirus (HPV)-dependent and -independent pathways, indicating a heterogeneous disease. Only a minority of VIN progress, but current clinicopathological classifications are insufficient to predict the cancer risk. Here we analyzed copy number alterations (CNA) to assess the molecular heterogeneity of vulvar lesions in relation to HPV and cancer risk. HPV-status and CNA by means of whole-genome next-generation shallow-sequencing were assessed in VSCC and VIN. The latter included VIN of women with associated VSCC (VINVSCC ) and women who did not develop VSCC during follow-up (VINnoVSCC ). HPV-testing resulted in 41 HPV-positive (16 VINVSCC , 14 VINnoVSCC , and 11 VSCC) and 24 HPV-negative (11 VINVSCC and 13 VSCC) lesions. HPV-positive and -negative VSCC showed a partially overlapping pattern of recurrent CNA, including frequent gains of 3q and 8q. In contrast, amplification of 11q13/cyclinD1 was exclusively found in HPV-negative lesions. HPV-negative VINVSCC had less CNA than HPV-negative VSCC (P = .009), but shared chromosome 8 alterations. HPV-positive VINnoVSCC had less CNA than VINVSCC (P = .022). Interestingly, 1pq gain was detected in 81% of HPV-positive VINVSCC and only in 21% of VINnoVSCC (P = .001). In conclusion, HPV-dependent and -independent vulvar carcinogenesis is characterized by distinct CNA patterns at the VIN stage, while more comparable patterns are present at the cancer stage. Cancer risk in VIN seems to be reflected by the extent of CNA, in particular chromosome 1 gain in HPV-positive cases.
Keywords: copy number alterations; human papillomavirus; progression risk; vulvar intraepithelial neoplasia; vulvar squamous cell carcinoma.
© 2018 The Authors. Cancer Medicine published by John Wiley & Sons Ltd.
Figures
References
-
- Trietsch MD, Nooij LS, Gaarenstroom KN, van Poelgeest MI. Genetic and epigenetic changes in vulvar squamous cell carcinoma and its precursor lesions: a review of the current literature. Gynecol Oncol. 2015;136:143‐157. - PubMed
-
- Del Pino M, Rodriguez‐Carunchio L, Ordi J. Pathways of vulvar intraepithelial neoplasia and squamous cell carcinoma. Histopathology. 2013;62:161‐175. - PubMed
-
- de Sanjose S, Alemany L, Ordi J, et al. Worldwide human papillomavirus genotype attribution in over 2000 cases of intraepithelial and invasive lesions of the vulva. Eur J Cancer. 2013;49:3450‐3461. - PubMed
-
- Bleeker MC, Visser PJ, Overbeek LI, van Beurden M, Berkhof J. Lichen sclerosus: incidence and risk of vulvar squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2016;25:1224‐1230. - PubMed
-
- Faber MT, Sand FL, Albieri V, Norrild B, Kjaer SK, Verdoodt F. Prevalence and type distribution of human papillomavirus in squamous cell carcinoma and intraepithelial neoplasia of the vulva. Int J Cancer. 2017;141:1161‐1169. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

