Single-Cell RNA Sequencing: A New Window into Cell Scale Dynamics
- PMID: 30033145
- PMCID: PMC6084636
- DOI: 10.1016/j.bpj.2018.07.003
Single-Cell RNA Sequencing: A New Window into Cell Scale Dynamics
Abstract
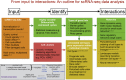
Single-cell genomics has recently emerged as a powerful tool for observing multicellular systems at a much higher level of resolution and depth than previously possible. High-throughput single-cell RNA sequencing techniques are able to simultaneously quantify expression levels of several thousands of genes within individual cells for tens of thousands of cells within a complex tissue. This has led to development of novel computational methods to analyze this high-dimensional data, investigating longstanding and fundamental questions regarding the granularity of cell types, the definition of cell states, and transitions from one cell type to another along developmental trajectories. In this perspective, we outline this emerging field starting from the "input data" (e.g., quantifying transcription levels in single cells), which are analyzed to define "identities" (e.g., cell types, states, and key genes) and to build "interactions" using models that can infer relations and transitions between cells.
Copyright © 2018 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Concepts and limitations for learning developmental trajectories from single cell genomics.Development. 2019 Jun 27;146(12):dev170506. doi: 10.1242/dev.170506. Development. 2019. PMID: 31249007 Review.
-
Single-cell cDNA microarray profiling of complex biological processes of differentiation.Curr Opin Genet Dev. 2010 Oct;20(5):470-7. doi: 10.1016/j.gde.2010.06.003. Epub 2010 Jul 7. Curr Opin Genet Dev. 2010. PMID: 20619631 Review.
-
Comparative Analysis of Droplet-Based Ultra-High-Throughput Single-Cell RNA-Seq Systems.Mol Cell. 2019 Jan 3;73(1):130-142.e5. doi: 10.1016/j.molcel.2018.10.020. Epub 2018 Nov 21. Mol Cell. 2019. PMID: 30472192
-
Spatiotemporal Developmental Trajectories in the Arabidopsis Root Revealed Using High-Throughput Single-Cell RNA Sequencing.Dev Cell. 2019 Mar 25;48(6):840-852.e5. doi: 10.1016/j.devcel.2019.02.022. Dev Cell. 2019. PMID: 30913408
-
Single-Cell RNA Sequencing of Oligodendrocyte Lineage Cells from the Mouse Central Nervous System.Methods Mol Biol. 2019;1936:1-21. doi: 10.1007/978-1-4939-9072-6_1. Methods Mol Biol. 2019. PMID: 30820890
Cited by
-
Statistical and Machine Learning Approaches to Predict Gene Regulatory Networks From Transcriptome Datasets.Front Plant Sci. 2018 Nov 29;9:1770. doi: 10.3389/fpls.2018.01770. eCollection 2018. Front Plant Sci. 2018. PMID: 30555503 Free PMC article. Review.
-
Building gene regulatory networks from scATAC-seq and scRNA-seq using Linked Self Organizing Maps.PLoS Comput Biol. 2019 Nov 4;15(11):e1006555. doi: 10.1371/journal.pcbi.1006555. eCollection 2019 Nov. PLoS Comput Biol. 2019. PMID: 31682608 Free PMC article.
-
Personalized medicine-concepts, technologies, and applications in inflammatory skin diseases.APMIS. 2019 May;127(5):386-424. doi: 10.1111/apm.12934. APMIS. 2019. PMID: 31124204 Free PMC article. Review.
-
A Method for Bioluminescence-Based RNA Monitoring Using Split-Luciferase Reconstitution Techniques.Methods Mol Biol. 2025;2875:9-20. doi: 10.1007/978-1-0716-4248-1_2. Methods Mol Biol. 2025. PMID: 39535636
-
EMBEDR: Distinguishing signal from noise in single-cell omics data.Patterns (N Y). 2022 Feb 8;3(3):100443. doi: 10.1016/j.patter.2022.100443. eCollection 2022 Mar 11. Patterns (N Y). 2022. PMID: 35510181 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

