Bacteriophage Cooperation Suppresses CRISPR-Cas3 and Cas9 Immunity
- PMID: 30033364
- PMCID: PMC6086726
- DOI: 10.1016/j.cell.2018.06.013
Bacteriophage Cooperation Suppresses CRISPR-Cas3 and Cas9 Immunity
Abstract
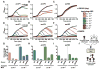
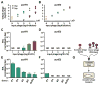
Bacteria utilize CRISPR-Cas adaptive immune systems for protection from bacteriophages (phages), and some phages produce anti-CRISPR (Acr) proteins that inhibit immune function. Despite thorough mechanistic and structural information for some Acr proteins, how they are deployed and utilized by a phage during infection is unknown. Here, we show that Acr production does not guarantee phage replication when faced with CRISPR-Cas immunity, but instead, infections fail when phage population numbers fall below a critical threshold. Infections succeed only if a sufficient Acr dose is contributed to a single cell by multiple phage genomes. The production of Acr proteins by phage genomes that fail to replicate leave the cell immunosuppressed, which predisposes the cell for successful infection by other phages in the population. This altruistic mechanism for CRISPR-Cas inhibition demonstrates inter-virus cooperation that may also manifest in other host-parasite interactions.
Keywords: CRISPR-Cas immunity; CRISPR-Cas9; Pseudomonas aeruginosa; altruism; anti-CRISPR; bacteriophage; cooperation; host-pathogen interaction; virus.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures


Comment in
-
Viruses cooperate to defeat bacteria.Nature. 2018 Jul;559(7715):482-484. doi: 10.1038/d41586-018-05762-1. Nature. 2018. PMID: 30038344 No abstract available.
-
Viral Teamwork Pushes CRISPR to the Breaking Point.Cell. 2018 Aug 9;174(4):772-774. doi: 10.1016/j.cell.2018.07.025. Cell. 2018. PMID: 30096306
-
Viral Anti-CRISPR Tactics: No Success without Sacrifice.Immunity. 2018 Sep 18;49(3):391-393. doi: 10.1016/j.immuni.2018.08.023. Immunity. 2018. PMID: 30231980
References
-
- Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050. - PubMed
-
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. - PubMed
-
- Bikard D, Hatoum-Aslan A, Mucida D, Marraffini LA. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe. 2012;12:177–186. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

