Anti-CRISPR Phages Cooperate to Overcome CRISPR-Cas Immunity
- PMID: 30033365
- PMCID: PMC6086933
- DOI: 10.1016/j.cell.2018.05.058
Anti-CRISPR Phages Cooperate to Overcome CRISPR-Cas Immunity
Abstract
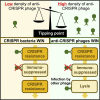
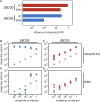
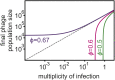
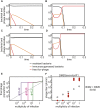
Some phages encode anti-CRISPR (acr) genes, which antagonize bacterial CRISPR-Cas immune systems by binding components of its machinery, but it is less clear how deployment of these acr genes impacts phage replication and epidemiology. Here, we demonstrate that bacteria with CRISPR-Cas resistance are still partially immune to Acr-encoding phage. As a consequence, Acr-phages often need to cooperate in order to overcome CRISPR resistance, with a first phage blocking the host CRISPR-Cas immune system to allow a second Acr-phage to successfully replicate. This cooperation leads to epidemiological tipping points in which the initial density of Acr-phage tips the balance from phage extinction to a phage epidemic. Furthermore, both higher levels of CRISPR-Cas immunity and weaker Acr activities shift the tipping points toward higher initial phage densities. Collectively, these data help elucidate how interactions between phage-encoded immune suppressors and the CRISPR systems they target shape bacteria-phage population dynamics.
Keywords: Allee effect; CRISPR-Cas; anti-CRISPR; bacteria; bifurcation; epidemiology; immunosuppression; partial resistance; phage; tipping points.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures







Comment in
-
Viruses cooperate to defeat bacteria.Nature. 2018 Jul;559(7715):482-484. doi: 10.1038/d41586-018-05762-1. Nature. 2018. PMID: 30038344 No abstract available.
-
Viral Teamwork Pushes CRISPR to the Breaking Point.Cell. 2018 Aug 9;174(4):772-774. doi: 10.1016/j.cell.2018.07.025. Cell. 2018. PMID: 30096306
-
Viral Anti-CRISPR Tactics: No Success without Sacrifice.Immunity. 2018 Sep 18;49(3):391-393. doi: 10.1016/j.immuni.2018.08.023. Immunity. 2018. PMID: 30231980
References
-
- Andersson A.F., Banfield J.F. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050. - PubMed
-
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

